BAUH: MND scRNA-seq pilot 2: dropkick
C.B. Azodi
2021-09-27
Last updated: 2021-09-27
Checks: 6 1
Knit directory: BAUH_2020_MND-single-cell/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190102) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e5d517e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/1-s2.0-S0002929720300781-main.pdf
Ignored: data/2103.11251.pdf
Ignored: data/3M-february-2018.txt
Ignored: data/737K-august-2016.txt
Ignored: data/STAR_index/
Ignored: data/STAR_output/
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.log
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.vcf.gz
Ignored: data/genome1k.chr22.log
Ignored: data/genome1k.chr22.recode.vcf
Ignored: data/s41588-018-0268-8.pdf
Ignored: data/tr2g_hs.tsv
Ignored: logs/
Ignored: output/CB-scRNAv31-GEX-lib01_QC_metadata.txt
Ignored: output/CB-scRNAv31-GEX-lib02_QC_metadata.txt
Ignored: output/pilot1_starsoloED/
Ignored: output/pilot2.1_gex/
Ignored: output/pilot2_HTO-2/
Ignored: output/pilot2_HTO/
Ignored: output/pilot2_gex_MAF01-152/
Ignored: output/pilot2_gex_MAF01/
Ignored: output/pilot2_gex_starsolo/
Ignored: output/pilot2_gex_starsoloED/
Ignored: output/pilot2_gex_starsoloED_GFP/
Ignored: references/SAindex/
Ignored: references/geno_test.vcf.gz
Untracked files:
Untracked: .snakemake/
Untracked: 2021-04-27_pilot2_nCells-per-donor.pdf
Untracked: 2021-08-03_pilot2_nCells-per-donor.pdf
Untracked: BAUH_2020_MND-single-cell.Rproj
Untracked: GRCh38_turboGFP-RFP_reference/
Untracked: Log.out
Untracked: Rplots.pdf
Untracked: analysis/2021-08-03_BAUH_MND-pilot-2.Rmd
Untracked: analysis/2021-09-27_pilot2_EmptyDrops.Rmd
Untracked: star-help.txt
Untracked: test/
Untracked: test_learn/
Untracked: test_maf01_notFiltered/
Untracked: test_maf05/
Untracked: test_noGeno/
Untracked: test_vireo/
Untracked: workflow/dropkick_get_ambient_and_hvgs.py
Unstaged changes:
Modified: analysis/2021-06-28_dropkick-results.Rmd
Modified: analysis/2021-07-07_geno-guided-donor-assignment-tests.Rmd
Modified: analysis/2021-07-07_geno-guided-donor-assignment-tests.html
Modified: analysis/index.Rmd
Modified: workflow/config_ambient_pilot2.1.yml
Staged changes:
Modified: analysis/2021-07-07_geno-guided-donor-assignment-tests.Rmd
New: analysis/2021-07-07_geno-guided-donor-assignment-tests.html
Modified: analysis/2021-08-04_DropletQC.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/2021-06-28_dropkick-results.Rmd) and HTML (public/2021-06-28_dropkick-results.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9fa3ac2 | cazodi | 2021-08-06 | dropkick figures |
| html | 9fa3ac2 | cazodi | 2021-08-06 | dropkick figures |
dropkick cellcalling
Dropkick is a fully automated software tool for quality control and filtering scRNA-seq data with a focus on excluding ambient barcodes and recovering only real cells. By automatically determining dataset-specific training labels based on predictive global heuristics, dropkick learns a gene-based representation of real cells and ambient noise, calculating a cell probability score for each barcode. Unlike EmptyDroplet, it does not set heuristic minimum thresholds for counts per cell.
Here is an example of dropkick results on a pan T-cell dataset from the manuscript:
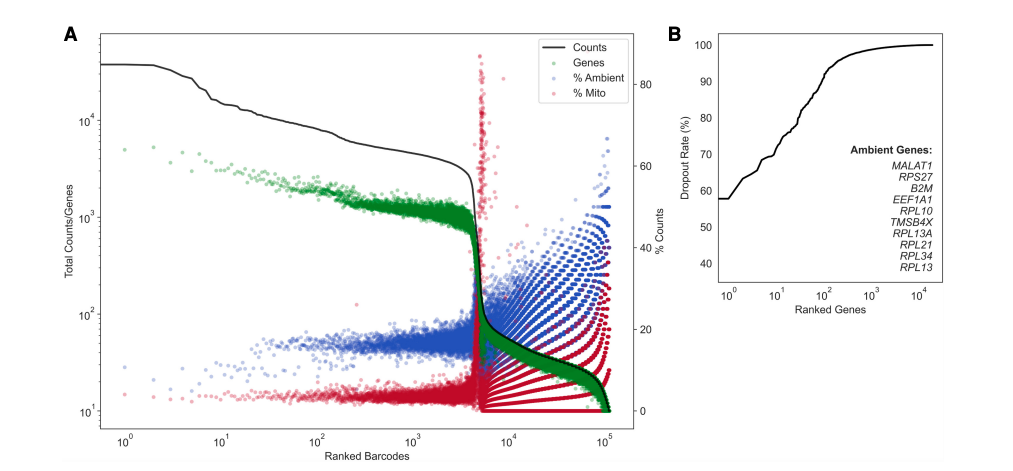
(left) a profile of total counts (black trace) and genes (green points) detected per ranked barcode, with the percentage of mitochondrial (red) and ambient (blue) reads for each barcode included to denote quality along dataset profile. (right) The dropout rate per gene ranked where top ranked genes are present in all barcodes (i.e. likely ambient). Ambient genes are identified by dropkick and used to calculate ambient percentage in the left figure.
Now here are the results from the pilot #2 data (unfixed):
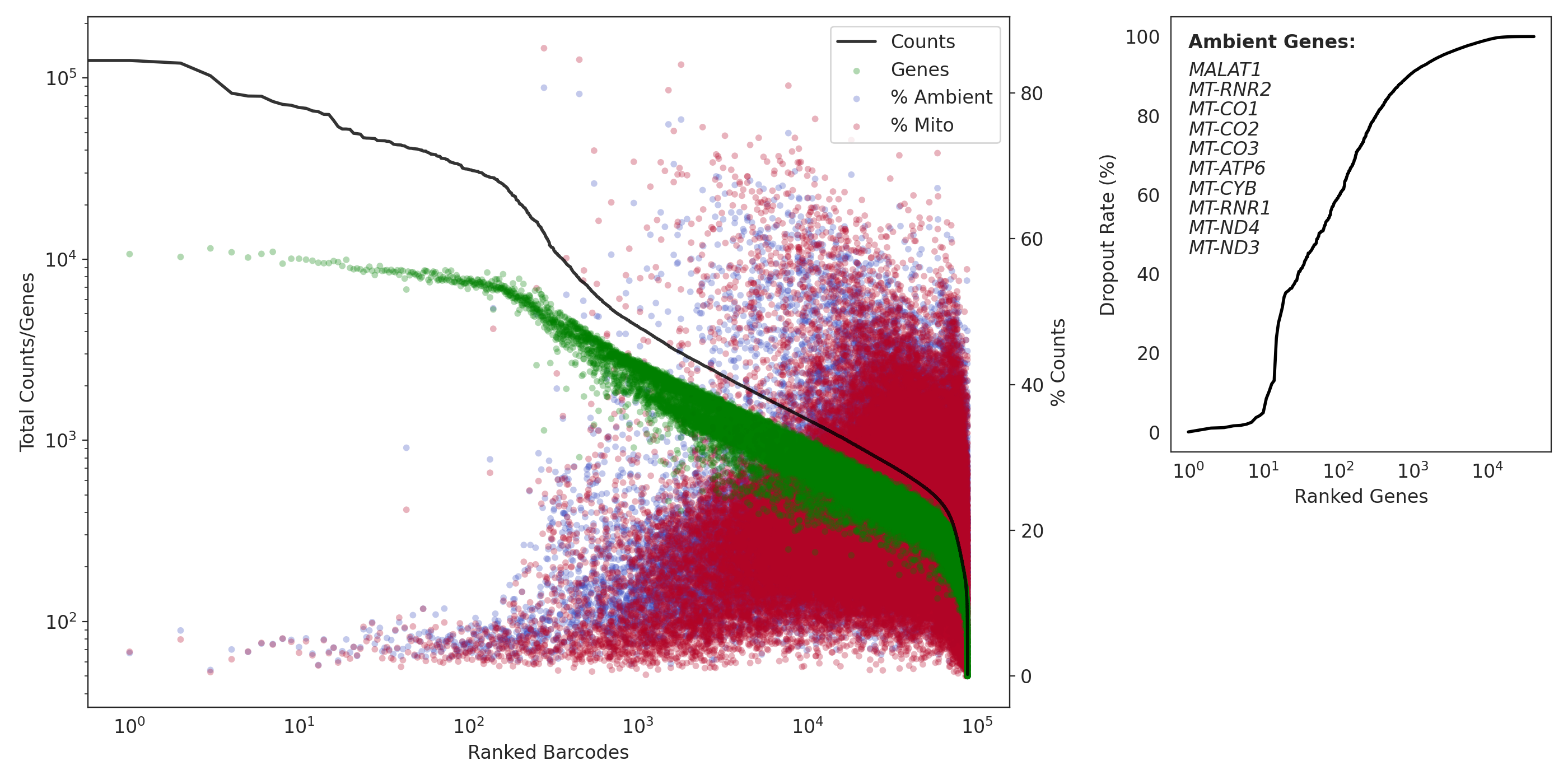
(same as above)
A major difference is the large number of genes in our data that have a 0% or close to 0% dropout rate. Genes with such low dropout are highly likely to be ambient noise! In our dataset, nearly the first 1000 genes have an dropout rate less than the most ambient gene in the example data!
dropkick results
Dropkick then learns a cell model and assigns each barcode a dropkick score, where a high score indicates a likely cell. The figure below shows the percent ambient counts versus arcsinh-transformed genes detected per barcode, with histogram distributions plotted on margins. Initial dropkick thresholds defining the training set are shown as dashed vertical lines. Each point (barcode) is colored by its final dropkick score after model fitting.
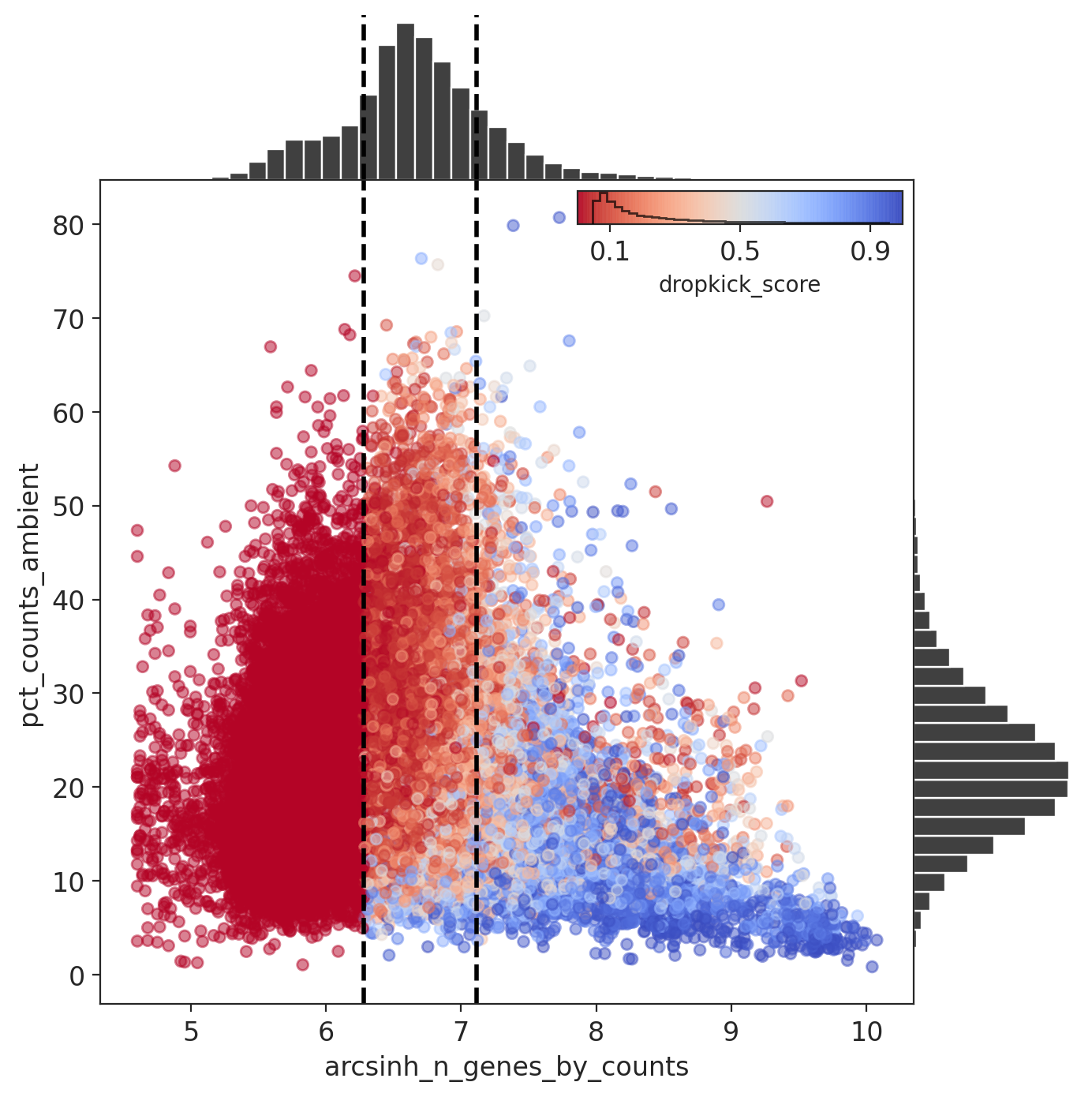
dropkick results
Compared to the example results from the manuscript below, we see that there is a less well defined boundary between cells and empty barcodes, which we expected based on the initial knee plots.
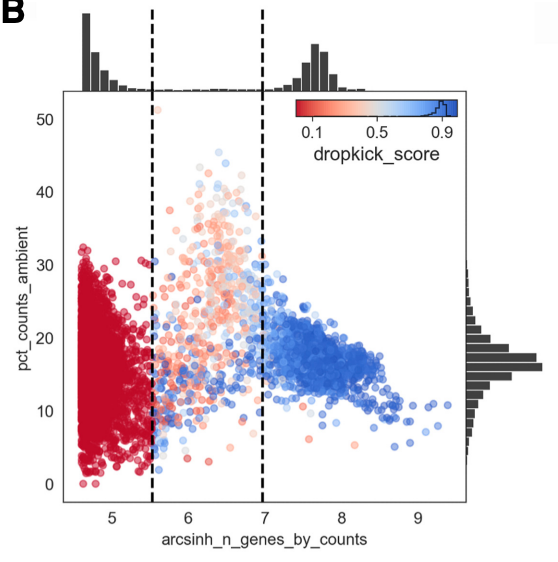
example dropkick results
Dropkick called 11,019 barcodes as cells from the unfixed experiment, nearly twice as many as called by EmptyDroplets (n=5,695)
We also ran dropkick on the fixed cells, which returned only 107 cells, even less than EmptyDroplet (n=609). Given we expected the fixed cells to be low quality, this seems promising.
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.0.4 (2021-02-15)
os Rocky Linux 8.4 (Green Obsidian)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-09-27
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
bslib 0.2.5.1 2021-05-18 [1] CRAN (R 4.0.4)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.4)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.0.4)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.0.4)
crayon 1.4.1 2021-02-08 [1] CRAN (R 4.0.4)
desc 1.3.0 2021-03-05 [1] CRAN (R 4.0.4)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.0.4)
digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.2)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.0.4)
evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.2)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.0.4)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.0.3)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.0.4)
glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.0.4)
httpuv 1.6.2 2021-08-18 [1] CRAN (R 4.0.4)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.4)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.4)
knitr 1.33 2021-04-24 [1] CRAN (R 4.0.4)
later 1.3.0 2021-08-18 [1] CRAN (R 4.0.4)
lifecycle 1.0.0 2021-02-15 [1] CRAN (R 4.0.4)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.3)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.4)
pillar 1.6.2 2021-07-29 [1] CRAN (R 4.0.4)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.4)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
pkgload 1.2.1 2021-04-06 [1] CRAN (R 4.0.4)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.2)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.0.4)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.0.4)
ps 1.6.0 2021-02-28 [1] CRAN (R 4.0.4)
purrr 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.0.4)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.0.4)
remotes 2.4.0 2021-06-02 [1] CRAN (R 4.0.4)
rlang 0.4.11 2021-04-30 [1] CRAN (R 4.0.4)
rmarkdown 2.10 2021-08-06 [1] CRAN (R 4.0.4)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.3)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.3)
sass 0.4.0 2021-05-12 [1] CRAN (R 4.0.4)
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
stringi 1.7.3 2021-07-16 [1] CRAN (R 4.0.4)
stringr 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
testthat 3.0.4 2021-07-01 [1] CRAN (R 4.0.4)
tibble 3.1.3 2021-07-23 [1] CRAN (R 4.0.4)
usethis 2.0.1 2021-02-10 [1] CRAN (R 4.0.4)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.4)
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.0.4)
whisker 0.4 2019-08-28 [1] CRAN (R 4.0.2)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.0.4)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.0.2)
xfun 0.25 2021-08-06 [1] CRAN (R 4.0.4)
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
[1] /mnt/mcfiles/cazodi/R/x86_64-pc-linux-gnu-library/4.0
[2] /opt/R/4.0.4/lib/R/library