2021-07-21_FANCM-crossovers-integrative-analysis
Ruqian Lyu
7/21/2021
Last updated: 2021-12-17
Checks: 6 1
Knit directory: yeln_2019_spermtyping/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190102) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 395a4f8. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .config/
Ignored: analysis/.Rhistory
Ignored: data/F21FTSAPHT0641MOUobtfR/
Untracked files:
Untracked: .DS_Store
Untracked: .Renviron
Untracked: .bulkDNAseq_BGI-extra.snk.swp
Untracked: .cache/
Untracked: .snakemake/
Untracked: .theano/
Untracked: 2021-07-13_scRNA-seq-analysis-of-sperm-cells-from-Fancm-mutant-and-Fancm-wildtype-mice.Rmd
Untracked: 2021-07-13_scRNA-seq-analysis-of-sperm-cells-from-Fancm-mutant-and-Fancm-wildtype-mice_files/
Untracked: 2021-09-10_BC1F1-copy-number-analysis.Rmd
Untracked: 2021-10-27_GvizDiagnostic.Rmd
Untracked: 2021-10-27_GvizDiagnostic.html
Untracked: 2021-10-27_GvizDiagnostic.log
Untracked: 2021-10-27_GvizDiagnostic.tex
Untracked: Rplots.pdf
Untracked: Snakefile_AFtracks.log.out
Untracked: Snakefile_BC1F1BGIseq2021.snk
Untracked: Snakefile_DNAseq-BGIseq-bc1f1-hmm-2021.log.out
Untracked: Snakefile_DNAseq-BGIseq-bc1f1-hmm-extra-11.log.out
Untracked: Snakefile_DNAseq-BGIseq-bcf-hmm.log.out
Untracked: Snakefile_DNAseq-F2F3-hmm.snk.log.out
Untracked: Snakefile_DNAseq-agrf-hmm.snk.log.out
Untracked: Snakefile_DNAseq-mos.log.out
Untracked: Snakefile_DNAseqF2F3.log.out
Untracked: Snakefile_alignATACseq.log.out
Untracked: Snakefile_bulkATACseq.snk
Untracked: Snakefile_bulkBGILowTrans.log.out
Untracked: Snakefile_bulkagrcountCO.log.out
Untracked: Snakefile_bulkagrf.log.out
Untracked: Snakefile_bulkagrfLowTrans.log.out
Untracked: Snakefile_bulkagrfQC.log.out
Untracked: Snakefile_bulkagrfSegRatio.log.out
Untracked: Snakefile_bulkagrffilterCO.log.out
Untracked: Snakefile_bulkbc1f1-2021.log.out
Untracked: Snakefile_bulkbgi.log.out
Untracked: Snakefile_bulkbgifilterCO.log.out
Untracked: Snakefile_cellChrSNP.err.log.out
Untracked: Snakefile_extraBulkbgi.log.out
Untracked: Snakefile_plotDiagnosticAFtracks.snk
Untracked: Snakefile_runCNVcaller.snk
Untracked: Snakefile_runGCNV.log.out
Untracked: Snakefile_runHMMCNV.log.out
Untracked: Snakefile_runcsnp-rnaseq.snk
Untracked: Snakefile_split.fq.out
Untracked: Snakefile_tag-fastq-bc.snk
Untracked: Snakefile_tag.err.log.out
Untracked: W12/
Untracked: WC_522/
Untracked: WC_526/
Untracked: WC_CNV_42/
Untracked: WC_CNV_42_firstBatch/
Untracked: WC_CNV_43/
Untracked: WC_CNV_43_firstBatch/
Untracked: WC_CNV_44/
Untracked: WC_CNV_44_firstBatch/
Untracked: WC_CNV_53/
Untracked: WC_CNV_53_firstBatch/
Untracked: WC_CNV_md5.txt
Untracked: WC_Sp/
Untracked: analysis/2021-01-14_lowCov-DNAseqAGRF-coverage.Rmd
Untracked: analysis/2021-07-13_scRNA-seq analysis of sperm cells from Fancm-mutant and Fancm-wildtype mice.Rmd
Untracked: analysis/2021-07-14_ATACseq-coverage.Rmd
Untracked: analysis/2021-07-26_BC1F1-SampleCNVs.Rmd
Untracked: analysis/2021-10-27_GvizDiagnostic.Rmd
Untracked: analysis/2021-11-07_BinnedGenDistComparison.Rmd
Untracked: analysis/2021-11-19_Permutation-testing-genetic-map-diferences-in-genotype-group.Rmd
Untracked: analysis/nstates.png
Untracked: bc1f1_chr19bin_state_gr.rds
Untracked: code/bc1f1-af-co-tracks.R
Untracked: code/binom-seg-test-scCNV.R
Untracked: code/chromosomeCODensityLinearRegression.R
Untracked: code/investigateSegsCNV.R
Untracked: code/run_sccomp.R
Untracked: code/scCNV_CO_SNP_density.R
Untracked: dag-scCNV.svg
Untracked: dag.svg
Untracked: data/10xCNV/
Untracked: data/737K-crdna-v1.txt
Untracked: data/All_data_May_to_August_2019.xlsx
Untracked: data/BC1F1-bgi-2021.txt
Untracked: data/BC1F1-bgi-extra_2021/
Untracked: data/BC1F1-bgi-extra_2021_samplemeta.txt
Untracked: data/BC1F1-bgi-fq-path_2021/
Untracked: data/F20FTSAPHT0350_MUSyfqR_samplemeta.txt
Untracked: data/F2f3_samplemeta.txt
Untracked: data/agrfBulkDNA/
Untracked: data/agrf_samplefqdir.txt
Untracked: data/agrf_samplemeta.txt
Untracked: data/atac_seq/
Untracked: data/cellSNP_W12-time2.txt/
Untracked: data/convertWIG2bed.sh
Untracked: data/convertWIG2bed_2.sh
Untracked: data/fancm_mice/
Untracked: data/fancm_tubule_blinded_counts.xlsx
Untracked: data/genLn.sh
Untracked: data/hg19.gc5Base.txt.gz
Untracked: data/ibuprofen_fancm_bl6_tubule_counts.xlsx
Untracked: data/mm10.100k.bed
Untracked: data/mm10.100k.meanGC.bed
Untracked: data/mm10.100k.meanGC.bed.bk
Untracked: data/mm10.100k.meanGC.chr1.bed
Untracked: data/mm10.100k.meanGC.chr3.bed
Untracked: data/mm10.100k.sorted.bed
Untracked: data/mm10.1m.bed
Untracked: data/mm10.1m.meanGC.bed
Untracked: data/mm10.1m.meanGC.chr1.bed
Untracked: data/mm10.1m.meanGC.chr10.bed
Untracked: data/mm10.1m.meanGC.chr11.bed
Untracked: data/mm10.1m.meanGC.chr12.bed
Untracked: data/mm10.1m.meanGC.chr13.bed
Untracked: data/mm10.1m.meanGC.chr14.bed
Untracked: data/mm10.1m.meanGC.chr15.bed
Untracked: data/mm10.1m.meanGC.chr16.bed
Untracked: data/mm10.1m.meanGC.chr17.bed
Untracked: data/mm10.1m.meanGC.chr18.bed
Untracked: data/mm10.1m.meanGC.chr19.bed
Untracked: data/mm10.1m.meanGC.chr2.bed
Untracked: data/mm10.1m.meanGC.chr3.bed
Untracked: data/mm10.1m.meanGC.chr4.bed
Untracked: data/mm10.1m.meanGC.chr5.bed
Untracked: data/mm10.1m.meanGC.chr6.bed
Untracked: data/mm10.1m.meanGC.chr7.bed
Untracked: data/mm10.1m.meanGC.chr8.bed
Untracked: data/mm10.1m.meanGC.chr9.bed
Untracked: data/mm10.1m.sorted.bed
Untracked: data/mm10.bed
Untracked: data/mm10.gc5Base.bed
Untracked: data/mm10.gc5Base.chr1.bed
Untracked: data/mm10.gc5Base.chr10.bed
Untracked: data/mm10.gc5Base.chr11.bed
Untracked: data/mm10.gc5Base.chr12.bed
Untracked: data/mm10.gc5Base.chr13.bed
Untracked: data/mm10.gc5Base.chr14.bed
Untracked: data/mm10.gc5Base.chr15.bed
Untracked: data/mm10.gc5Base.chr16.bed
Untracked: data/mm10.gc5Base.chr17.bed
Untracked: data/mm10.gc5Base.chr18.bed
Untracked: data/mm10.gc5Base.chr19.bed
Untracked: data/mm10.gc5Base.chr2.bed
Untracked: data/mm10.gc5Base.chr3.bed
Untracked: data/mm10.gc5Base.chr4.bed
Untracked: data/mm10.gc5Base.chr5.bed
Untracked: data/mm10.gc5Base.chr6.bed
Untracked: data/mm10.gc5Base.chr7.bed
Untracked: data/mm10.gc5Base.chr8.bed
Untracked: data/mm10.gc5Base.chr9.bed
Untracked: data/mm10.gc5Base.wib
Untracked: data/mm10.gc5Base.wig.gz
Untracked: data/mm10.gc5Base.wigVarStep.gz
Untracked: data/public_dataset/
Untracked: data/rawNames.txt
Untracked: data/slurm-59654.out
Untracked: data/slurm-59655.out
Untracked: data/slurm-59670.out
Untracked: data/slurm-82444.out
Untracked: data/slurm-82445.out
Untracked: docs/figure/2020-10-20_BulkDNASamples-CO-genetic-dist.Rmd/
Untracked: docs/site_libs/accessible-code-block-0.0.1/
Untracked: envs/demultiplex.yml
Untracked: envs/env_bulkDNA.yml
Untracked: envs/mosdepth.yaml
Untracked: fastp.html
Untracked: fastp.json
Untracked: job_tracking/
Untracked: multipage.pdf
Untracked: myplot.png
Untracked: nohup.out
Untracked: nstates.png
Untracked: nstates1.png
Untracked: old_Renv
Untracked: output/.DS_Store
Untracked: output/10xCNV/
Untracked: output/atac_seq/
Untracked: output/bcf/
Untracked: output/bulkDNAseq/
Untracked: output/bulkDNAseq_BC1F1BGI2021/
Untracked: output/bulkDNAseq_BGI/
Untracked: output/bulkDNAseq_F2F3/
Untracked: output/bulkDNAseq_agrf/
Untracked: output/figures/
Untracked: output/gCNV/
Untracked: output/outputR/
Untracked: output/scCNV/
Untracked: output/scRNA/
Untracked: references/BC1F1_samples-2020-07-08.xlsx
Untracked: references/BC1F1_samples-2020-10-20.xlsx
Untracked: references/BC1F1_summary.xlsx
Untracked: references/Copy of crossover_count.csv
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.WC_522_het.vcf.gz
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.WC_522_het.vcf.gz.csi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.WC_522_het.vcf.gz.tbi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.WC_522_het.pos.only.tsv
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.WC_522_het.vcf.gz
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.WC_522_het.vcf.gz.csi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.WC_522_het.vcf.gz.tbi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.txt
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.vcf.gz
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.vcf.gz.csi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.chr.vcf.stat
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.vcf.gz
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.alt.vcf.gz.csi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.var.vcf
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.var.vcf.gz
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.var.vcf.gz.csi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.var.vcf.gz.tbi
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.var.vcf.idx
Untracked: references/FVB_NJ.mgp.v5.snps.dbSNP142.homo.var.vcf.stat
Untracked: references/cellranger-ref-dna-mm10/
Untracked: references/cellranger-ref-mm10/
Untracked: references/chrs.map
Untracked: references/chrs_r.map
Untracked: references/col1
Untracked: references/col2
Untracked: references/k36.umap.bed
Untracked: references/k36.umap.bed.idx
Untracked: references/mm10_contig_ploidy_priors.tsv
Untracked: references/scRNAseq_gene_sets.xlsx
Untracked: references/wasp/
Untracked: retag-0-mis.log.txt
Untracked: runCountCB.out
Untracked: run_countcb.snk
Untracked: runsCNV.out
Untracked: runsCsnp.out
Untracked: split.log.txt
Untracked: sslurm-42367.out
Untracked: submit-AlignBulkATAC.sh
Untracked: submit-runAlignBC1F1BGI.sh
Untracked: submit-runBulkDNAagrf.sh
Untracked: submit-runBulkDNAbgi.sh
Untracked: submit-runBulkDNAbgiExtra.sh
Untracked: submit-runCNV.sh
Untracked: submit-runCTCB.sh
Untracked: submit-runDNAseq-BGIseq-bc1f1-hmm-2021.sh
Untracked: submit-runDNAseq-BGIseq-bc1f1-hmm-Extra-2021.sh
Untracked: submit-runDNAseq-BGIseq-bcf-hmm.sh
Untracked: submit-runDNAseqF2F3.sh
Untracked: submit-runHMMBulkAGRF.sh
Untracked: submit-runHMMBulkAGRFBCF.sh
Untracked: submit-runHMMF2F3.sh
Untracked: submit-runMos.sh
Untracked: submit-runPlotAFBC1F1.sh
Untracked: submit-runSNK-cellchrsnp.sh
Untracked: submit-runSNK-hmm-CNV.sh
Untracked: submit-runcsnp.sh
Untracked: submit-rungCNV.sh
Untracked: submit-sbatch.sh
Untracked: test.png
Untracked: theano_base/
Untracked: tmp/
Untracked: yeln_2019_spermtyping.Rproj
Unstaged changes:
Modified: .gitignore
Modified: CITATION
Modified: LICENSE
Modified: Snakefile_DNAseq-BGIseq-bcf-hmm.snk
Modified: Snakefile_DNAseq-agrf-hmm.snk
Modified: Snakefile_DNAseq-chr-plot.snk
Modified: Snakefile_HMM-scCNV
Modified: Snakefile_bcf
Modified: Snakefile_bulkDNAseq-mos.snk
Modified: Snakefile_bulkDNAseq.snk
Modified: Snakefile_cluster
Modified: Snakefile_gatk
Modified: Snakefile_remap
Modified: _workflowr.yml
Modified: analysis/2019-04-13_expl-data-analysis_2019-04-12-10x-run.Rmd
Modified: analysis/2020-01-02_W12_smcelss_C9_bcftools.Rmd
Modified: analysis/2020-01-03_W11-small-cells.Rmd
Modified: analysis/2020-01-03_W11-vartrix-diploid.Rmd
Modified: analysis/2020-01-03_W11-vartrix.Rmd
Modified: analysis/2020-01-03_W11_get_smcells.Rmd
Modified: analysis/2020-01-03_W12-vartrix.Rmd
Modified: analysis/2020-05-01_10xCNV-summary.Rmd
Modified: analysis/2020-05-07_WC-522-bulk-sperm-vcf.Rmd
Modified: analysis/2020-05-11_WC-522-filtered-variants-single-cell.Rmd
Modified: analysis/2020-05-15_WC_522-wasp-Filtering.Rmd
Modified: analysis/2020-05-15_barcode-calling-10xCNV.Rmd
Modified: analysis/2020-05-19_SNP-gaps-and-distribution-analysis.Rmd
Modified: analysis/2020-05-25_FVB-BL6-Control-cell-for-estimating-HMM-pars.Rmd
Modified: analysis/2020-06-02_WC-522-nocell-group5.Rmd
Modified: analysis/2020-06-02_mix-haploid-cells.Rmd
Modified: analysis/2020-06-15_lowCov-DNAseq-summary-stats.Rmd
Modified: analysis/2020-06-16_detect-CO-bulkDNAseq-1X.Rmd
Modified: analysis/2020-06-17_scDNA-viterbi-CO-detection.Rmd
Modified: analysis/2020-07-06_bulk-samples-theta-estimate.Rmd
Modified: analysis/2020-07-14_agrf-genotyping-markers.Rmd
Modified: analysis/2020-07-27_state-bias-diagnosis.Rmd
Modified: analysis/2020-08-13_Marker-based-genetic-map.Rmd
Modified: analysis/2020-08-14_Genetic-map-hot-spots-bulk-BGI.Rmd
Modified: analysis/2020-08-18_total-genetic-dist-genome.Rmd
Modified: analysis/2020-10-06_scCNV-CO-genetic-dist.Rmd
Modified: analysis/2020-10-20_BulkDNASamples-CO-genetic-dist.Rmd
Modified: analysis/2020-10-20_agrfBulkDNASamples-WT-HET-CO-genetic-dist.Rmd
Modified: analysis/2020-11-24_BulkSamples-CrossoverAnalysisUpdates.Rmd
Modified: analysis/2020-7-22_detectCO-BulkDNA-corrected.Rmd
Modified: analysis/2021-02-09_F2F3-samples-CO-analysis.Rmd
Modified: analysis/2021-02-22_Investigation-Female-HET.Rmd
Modified: analysis/2021-04-05_GIMs-variants.Rmd
Modified: analysis/2021-06-28_BulkBC1F1-samples.Rmd
Modified: analysis/2021-06-29_exploring-data-batches-bulk-bc1f1-samples.Rmd
Modified: analysis/2021-07-05_Filters-Exploration-Bulk-BC1F1.Rmd
Modified: analysis/2021-07-21_FANCM-crossovers-integrative-analysis.Rmd
Modified: analysis/2021-07-23_FANCM-manuscript-figures-2.Rmd
Modified: analysis/2021-07-23_FANCM-manuscript-figures.Rmd
Modified: analysis/2021-07-25_FANCM-manuscript-figures-3.Rmd
Modified: analysis/2021-10-11_sccomp-compositional-testing-fancm-tubules.Rmd
Modified: analysis/2021-10-13_FANCM-manuscript-Crossovers-genomic-features.Rmd
Modified: analysis/2021-10-24_FancmCrossoverOutlierChromosome.Rmd
Modified: analysis/2021-11-01_hotspots-Fancm.Rmd
Modified: analysis/SNPs_hp_dp_cells.pptx
Modified: analysis/W11_W12_pooled_clustering.Rmd
Modified: analysis/W11_clustering.Rmd
Modified: analysis/W11_further_analysis.Rmd
Modified: analysis/W12_clustering.Rmd
Modified: analysis/about.Rmd
Modified: analysis/expl_W12_C28_GATK_AF_plot.Rmd
Modified: analysis/expl_W12_C28_cellsnp_AF_plot.Rmd
Modified: analysis/expl_W12_C2_cellsnp_AF_plot.Rmd
Modified: analysis/expl_W12_GATK_AF_plot.Rmd
Modified: analysis/expl_W12_bcftools_AF_plot.Rmd
Modified: analysis/expl_W12_bcftools_AF_plot_dp.Rmd
Modified: analysis/expl_W12_cellsnp_AF_plot.Rmd
Modified: analysis/license.Rmd
Modified: analysis/scRNAseq-analysis-outline.Rmd
Modified: bulkDNAseq_BGI-30X.snk
Modified: code/README.md
Modified: code/badSampleBulkDNAseq.R
Modified: code/bulk_BL6_FVB_rate0_estimate.R
Modified: code/cal_ref_ratio.R
Modified: code/cc_score.R
Modified: code/cellCO-QCControl.R
Modified: code/cellQualityCheck.R
Modified: code/cellQualityCheck.old.R
Modified: code/checkCoverageBulkseq.R
Modified: code/checkF2F3trios.R
Modified: code/checkF2F3triosLtrans.R
Modified: code/check_co.R
Modified: code/cluster.R
Modified: code/combine_calqc.R
Modified: code/combine_cluster.R
Modified: code/combine_crt_sce.R
Modified: code/combine_filter_qcdb.R
Modified: code/combine_findmarker.R
Modified: code/combine_monocle.R
Modified: code/combine_pca.R
Modified: code/combine_sctransnorm.R
Modified: code/combine_vis.R
Modified: code/controlCell-fvbbl6.R
Modified: code/controlCell-pEstimate.R
Modified: code/correctLowConfCO.R
Modified: code/count_cb.nim
Modified: code/cumGeneticDistance.R
Modified: code/def_viterbi.R
Modified: code/detect_noisy_cells.R
Modified: code/dr_pca.R
Modified: code/expl_bined_Ave.R
Modified: code/fancm_reads.R
Modified: code/filterStateAndCO.R
Modified: code/findLowConfCO.R
Modified: code/find_segment_length.R
Modified: code/findmarker.R
Modified: code/getMarkerBasedCOs.R
Modified: code/impute-chr-bin-state-bc1f1.R
Modified: code/markerStateAggr.R
Modified: code/nohup.out
Modified: code/norm.R
Modified: code/overlay_fun.R
Modified: code/plotBadMarkersBulk.R
Modified: code/plotCOregion.R
Modified: code/plotGeneticMap.R
Modified: code/plotHotRegion.R
Modified: code/plot_bp_bin-AF.R
Modified: code/plot_chr_co_bulkDNA.R
Modified: code/plot_corrected_state.R
Modified: code/plot_roh.py
Modified: code/rate_estimator.R
Modified: code/renamer.py
Modified: code/renamer_bam.nim
Modified: code/report_summary.Rmd
Modified: code/retag.py
Modified: code/retag_bam.nim
Modified: code/runVb-scCNV.R
Modified: code/run_viterbi.R
Modified: code/run_viterbi_F2F3.R
Modified: code/run_viterbi_bulkSample_chr.R
Modified: code/run_viterbi_cell_chr.R
Modified: code/scCNVcountCO.R
Modified: code/split_bam.nim
Modified: code/subsampleAGRF.R
Modified: code/vartrix.sh
Modified: code/viterbi_example_chr_cell.R
Modified: data/README.md
Modified: docs/.nojekyll
Modified: docs/2020-01-03_W11-small-cells.html
Modified: docs/Progress_report_yeln.html
Modified: docs/W11_W12_pooled_clustering.html
Modified: docs/W11_clustering.html
Modified: docs/W11_further_analysis.html
Modified: docs/W12_clustering.html
Modified: docs/W12_further_analysis.html
Modified: docs/about.html
Modified: docs/assets/F1_gametes.PNG
Modified: docs/assets/SplitNCigarReads.png
Modified: docs/assets/W11double.pdf
Modified: docs/assets/W12double.pdf
Modified: docs/assets/singleCell3pv2.PNG
Modified: docs/expl_W12_C28_GATK_AF_plot.html
Modified: docs/expl_W12_C28_cellsnp_AF_plot.html
Modified: docs/expl_W12_C2_cellsnp_AF_plot.html
Modified: docs/expl_W12_GATK_AF_plot.html
Modified: docs/expl_W12_bcftools_AF_plot.html
Modified: docs/expl_W12_bcftools_AF_plot_dp.html
Modified: docs/expl_W12_cellsnp_AF_plot.html
Modified: docs/expl_bcftools_dp_dedb_W12.html
Modified: docs/figure/Progress_report_yeln.Rmd/pressure-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_cluster-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_cluster-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_cluster-3.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_cluster-4.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_sample-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_sample-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/by_sample-3.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/c4_c28-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/cluster_composition-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/clustree-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/hm-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-10.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-11.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-12.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-13.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-14.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-15.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-16.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-3.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-4.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-5.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-6.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-7.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-8.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/known_markers-9.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster-3.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster-4.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster-5.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster-6.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster_by_sample-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_cluster_by_sample-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_fltqc-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_fltqc-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_hvg-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_marker_plot-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_pc_select-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_pca_visulisation-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_raw_qc-1.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/pulled_raw_qc-2.png
Modified: docs/figure/W11_W12_pooled_clustering.Rmd/unnamed-chunk-4-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_cluster-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_cluster-2.png
Modified: docs/figure/W12_clustering.Rmd/W12_cluster-3.png
Modified: docs/figure/W12_clustering.Rmd/W12_cluster-4.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-10.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-11.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-12.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-13.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-14.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-15.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-2.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-3.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-4.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-5.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-6.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-7.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-8.png
Modified: docs/figure/W12_clustering.Rmd/W12_dedb_umaps-9.png
Modified: docs/figure/W12_clustering.Rmd/W12_fltqc-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_fltqc-2.png
Modified: docs/figure/W12_clustering.Rmd/W12_hvg-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_marker_plot-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_pc_select-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_pca_visulisation-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_raw_qc-1.png
Modified: docs/figure/W12_clustering.Rmd/W12_raw_qc-2.png
Modified: docs/figure/W12_clustering.Rmd/ab_genes-1.png
Modified: docs/figure/W12_clustering.Rmd/ab_genes-2.png
Modified: docs/figure/W12_clustering.Rmd/boxplot_chrYgenes-1.png
Modified: docs/figure/W12_clustering.Rmd/plotExpr-1.png
Modified: docs/figure/W12_clustering.Rmd/plotExpr-2.png
Modified: docs/figure/W12_clustering.Rmd/plotExpr-3.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-1.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-10.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-11.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-12.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-13.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-14.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-15.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-2.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-3.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-4.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-5.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-6.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-7.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-8.png
Modified: docs/figure/W12_clustering.Rmd/spermatids_markers-9.png
Modified: docs/figure/W12_clustering.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/W12_clustering.Rmd/unnamed-chunk-3-1.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/genotyped_AF-1.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/genotyped_AF-2.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/no_snps-2.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/unnamed-chunk-3-1.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/unnamed-chunk-3-2.png
Modified: docs/figure/expl_W12_C28_GATK_AF_plot.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/colr_dp-1.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/no_snps-2.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/no_snps-3.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/no_snps-4.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/no_snps-5.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/no_snps-6.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/rm_all_hets-1.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/unnamed-chunk-2-2.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/unnamed-chunk-4-1.png
Modified: docs/figure/expl_W12_C28_cellsnp_AF_plot.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/no_snps-2.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/no_snps-3.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/no_snps-4.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/no_snps-5.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/no_snps-6.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/rm_all_hets-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/rm_all_hets_dp-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/unnamed-chunk-3-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/unnamed-chunk-3-2.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/unnamed-chunk-5-1.png
Modified: docs/figure/expl_W12_C2_cellsnp_AF_plot.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/genotyped_AF-1.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/genotyped_AF-2.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/no_snps-2.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/unnamed-chunk-3-1.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/unnamed-chunk-3-2.png
Modified: docs/figure/expl_W12_GATK_AF_plot.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/all_bcf_cells-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/all_bcf_cells-2.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/colr_snp_dp-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/filter_qual-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/filter_qual-2.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/genotyped-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/genotyped-2.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/colr_snp_dp-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/filter_qual-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/filter_qual-2.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/genotyped-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/genotyped-2.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_bcftools_AF_plot_dp.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/filterSNPs-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/filterSNPs-2.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/no_snps-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/no_snps-2.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/no_snps-3.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/no_snps-4.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/no_snps-5.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/no_snps-6.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/unnamed-chunk-1-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/unnamed-chunk-4-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/unnamed-chunk-5-1.png
Modified: docs/figure/expl_W12_cellsnp_AF_plot.Rmd/whole_genome_af-1.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/genotyped-1.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/genotyped-2.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/no_snps-1.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/unnamed-chunk-1-1.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/unnamed-chunk-2-1.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/unnamed-chunk-2-2.png
Modified: docs/figure/expl_bcftools_dp_dedb_W12.Rmd/whole_genome_af-1.png
Modified: docs/index.html
Modified: docs/license.html
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cerulean.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cosmo.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/darkly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/flatly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Lato.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycle.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycleBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSans.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBoldItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLightItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Raleway.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RalewayBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Roboto.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoMedium.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansPro.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Ubuntu.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/journal.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/lumen.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/paper.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/readable.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/sandstone.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/simplex.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/spacelab.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/united.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/yeti.min.css
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.eot
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.svg
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.ttf
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff2
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.js
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.min.js
Modified: docs/site_libs/bootstrap-3.3.5/js/npm.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/html5shiv.min.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/respond.min.js
Modified: docs/site_libs/font-awesome-5.1.0/css/all.css
Modified: docs/site_libs/font-awesome-5.1.0/css/v4-shims.css
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff2
Modified: docs/site_libs/jquery-1.11.3/jquery.min.js
Modified: docs/site_libs/jqueryui-1.11.4/README
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_444444_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_555555_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777620_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777777_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_cc0000_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_ffffff_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/index.html
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.min.css
Modified: docs/site_libs/navigation-1.1/codefolding.js
Modified: docs/site_libs/navigation-1.1/sourceembed.js
Modified: docs/site_libs/navigation-1.1/tabsets.js
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.css
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.js
Modified: org/README.md
Modified: output/README.md
Modified: references/README.md
Modified: resources/README.md
Modified: yeln.bib
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/2021-07-21_FANCM-crossovers-integrative-analysis.Rmd) and HTML (public/2021-07-21_FANCM-crossovers-integrative-analysis.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 01458d7 | rlyu | 2021-10-01 | update integration analysis |
| html | 01458d7 | rlyu | 2021-10-01 | update integration analysis |
| Rmd | 9a3ae70 | rlyu | 2021-08-05 | adding segregation analysis results |
| html | 9a3ae70 | rlyu | 2021-08-05 | adding segregation analysis results |
Marker segregation
Test segregation bias for gametes.
[R scripts for preparing inputs for this report at: code/impute-chr-bin-state-scCNV.R]
Per sample: WC_522
This is done by taking the crossover results and work backwards to find the state for chromosome bins so that missing markers’ states are imputed. This helps to reduce the unreliable results for staring and ending bins of chromosomes.
Chr1
sampleName <- "WC_522"
chrName <- "chr1"
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))The chromosomes have been binned using size of 1e7.
bin_state_gr[1:4,1:3]GRanges object with 4 ranges and 3 metadata columns:
seqnames ranges strand | TGCGGGTGTATTGAAG-1 GTGCGGTGTGCTATTG-1
<Rle> <IRanges> <Rle> | <character> <character>
[1] chr1 1-9970630 * | s2 s1
[2] chr1 9970631-19941259 * | s2 s1
[3] chr1 19941260-29911888 * | s2 s1
[4] chr1 29911889-39882517 * | s2 s1
ATCTACTTCTAGCCTC-1
<character>
[1] s2
[2] s2
[3] s2
[4] s2
-------
seqinfo: 19 sequences from an unspecified genomedf <- mcols(bin_state_gr)
data.frame(df,check.names = FALSE) %>% dplyr::mutate(bin_id = seq(nrow(df))) %>%
tidyr::pivot_longer(cols = colnames(mcols(bin_state_gr))) %>%
ggplot()+geom_point(aes(y = bin_id, x = name,color = value))+theme_bw()+
theme(axis.text.x = element_blank())Test for any imbalance
Use binomial.test for testing whether proportion is 0.5
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",stat = "identity")+
geom_text(aes(x = bin_id,y=70, label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)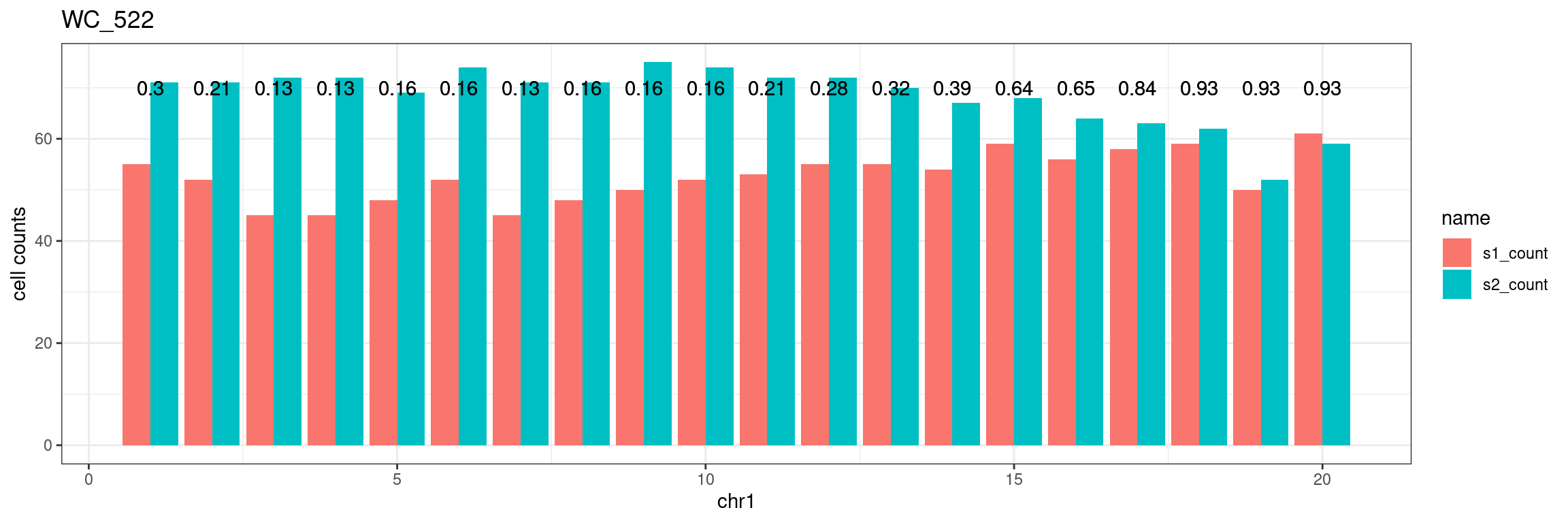
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
Produce the plots for all chrs
sampleName <- "WC_522"
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
stat = "identity")+
geom_text(aes(x = bin_id,y=70, label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)
plots_list[[chrName]] <- p+guides(fill = "none" )
} mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
mChrThresPlots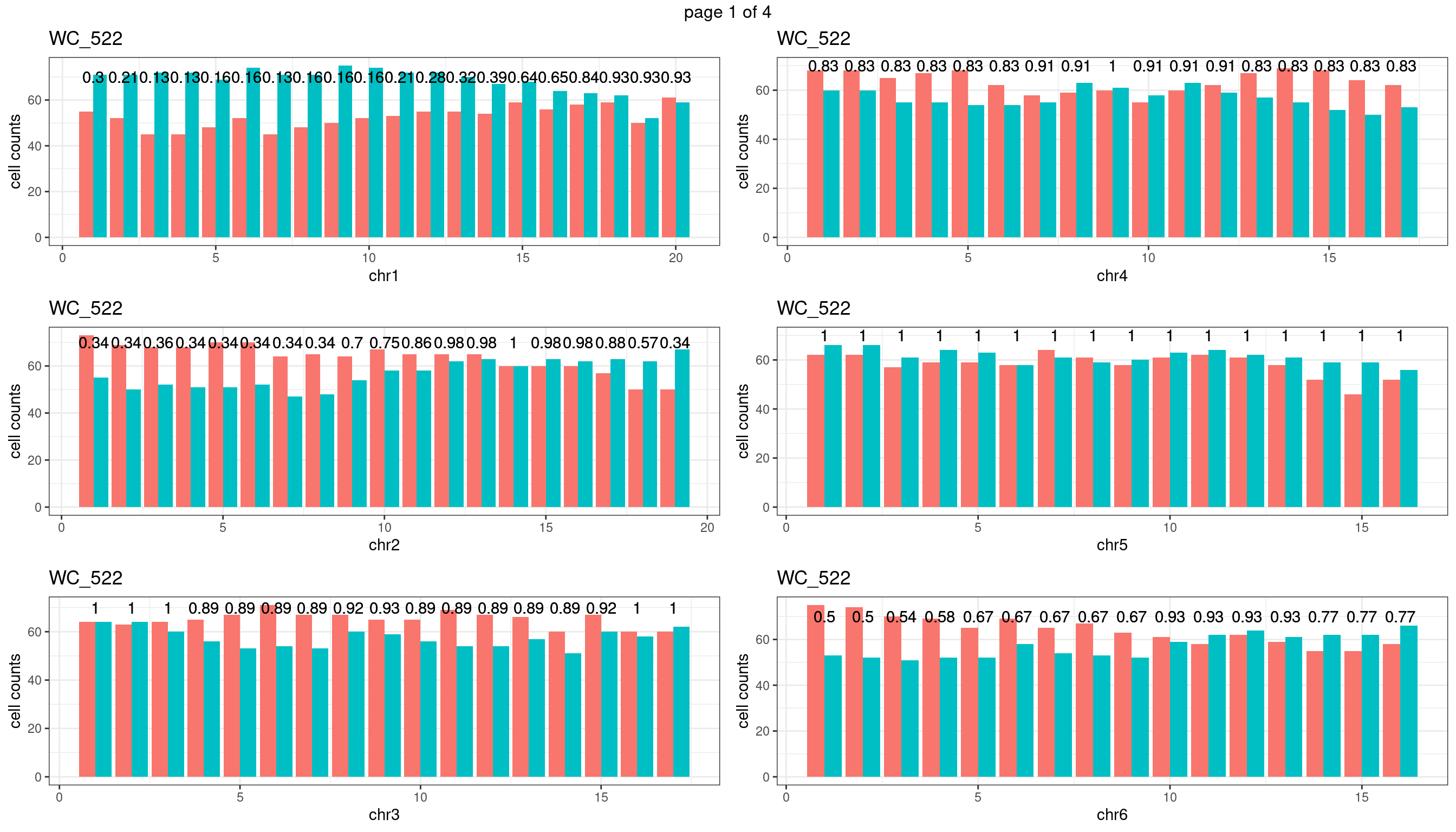
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
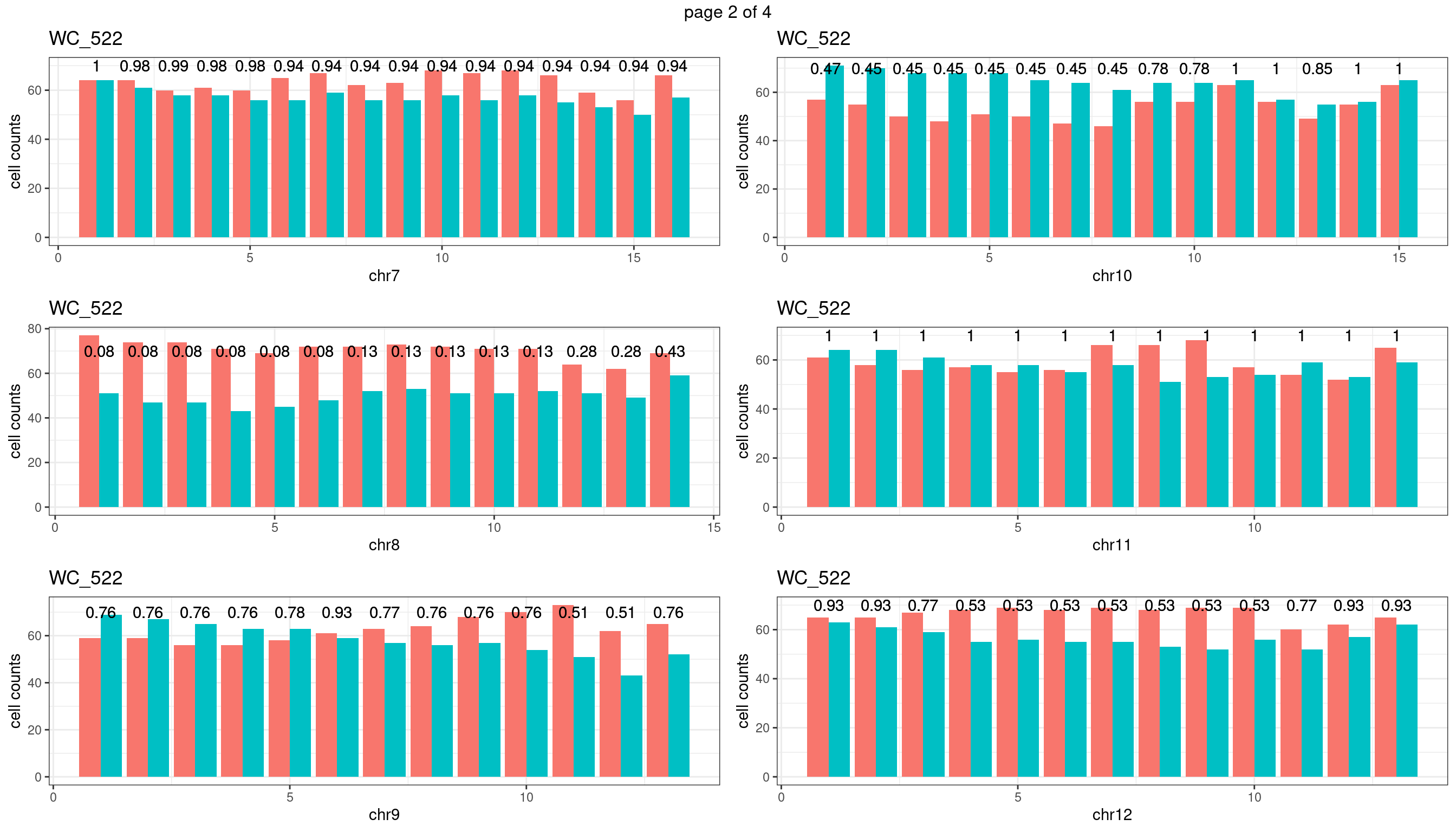
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
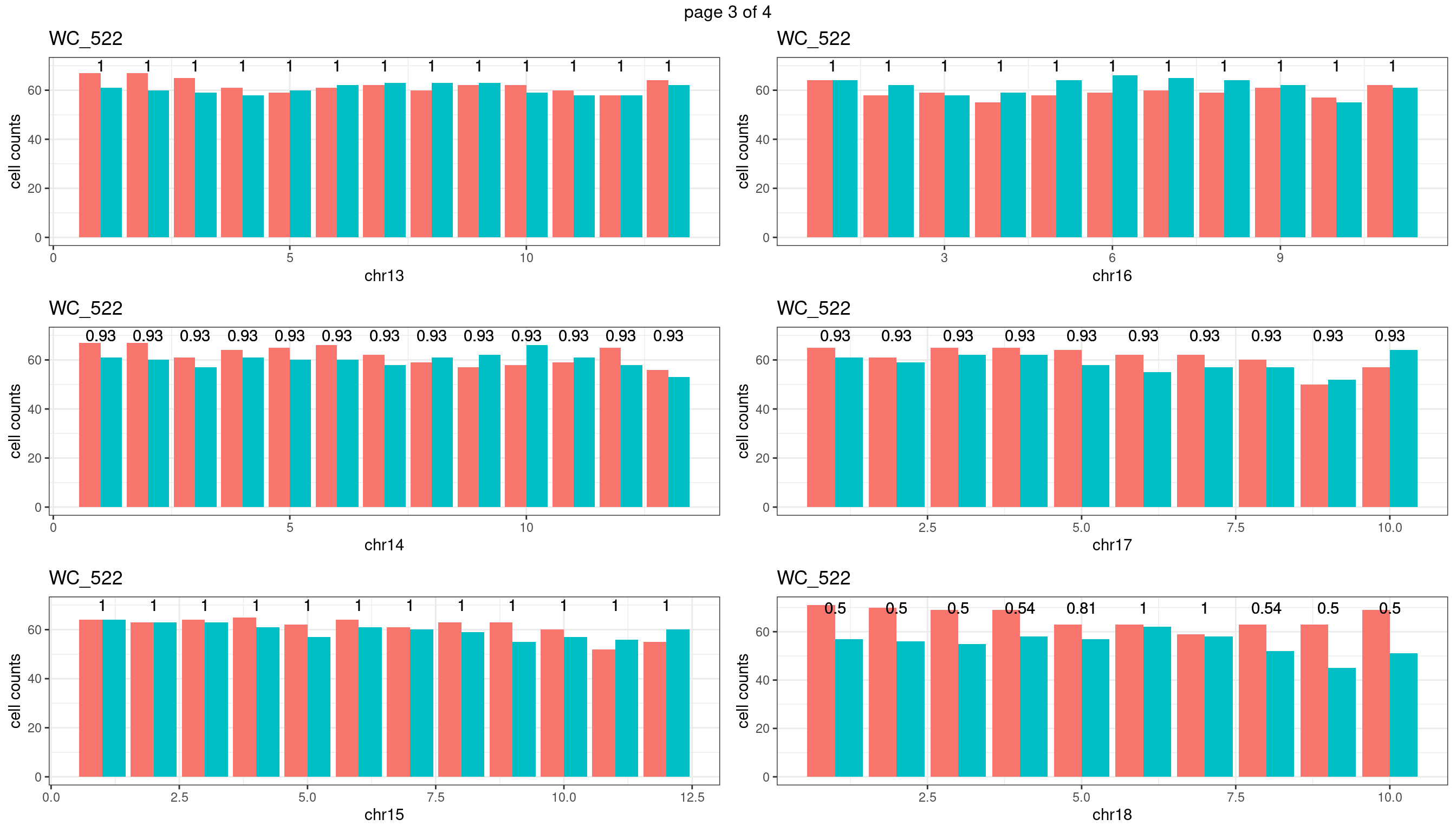
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
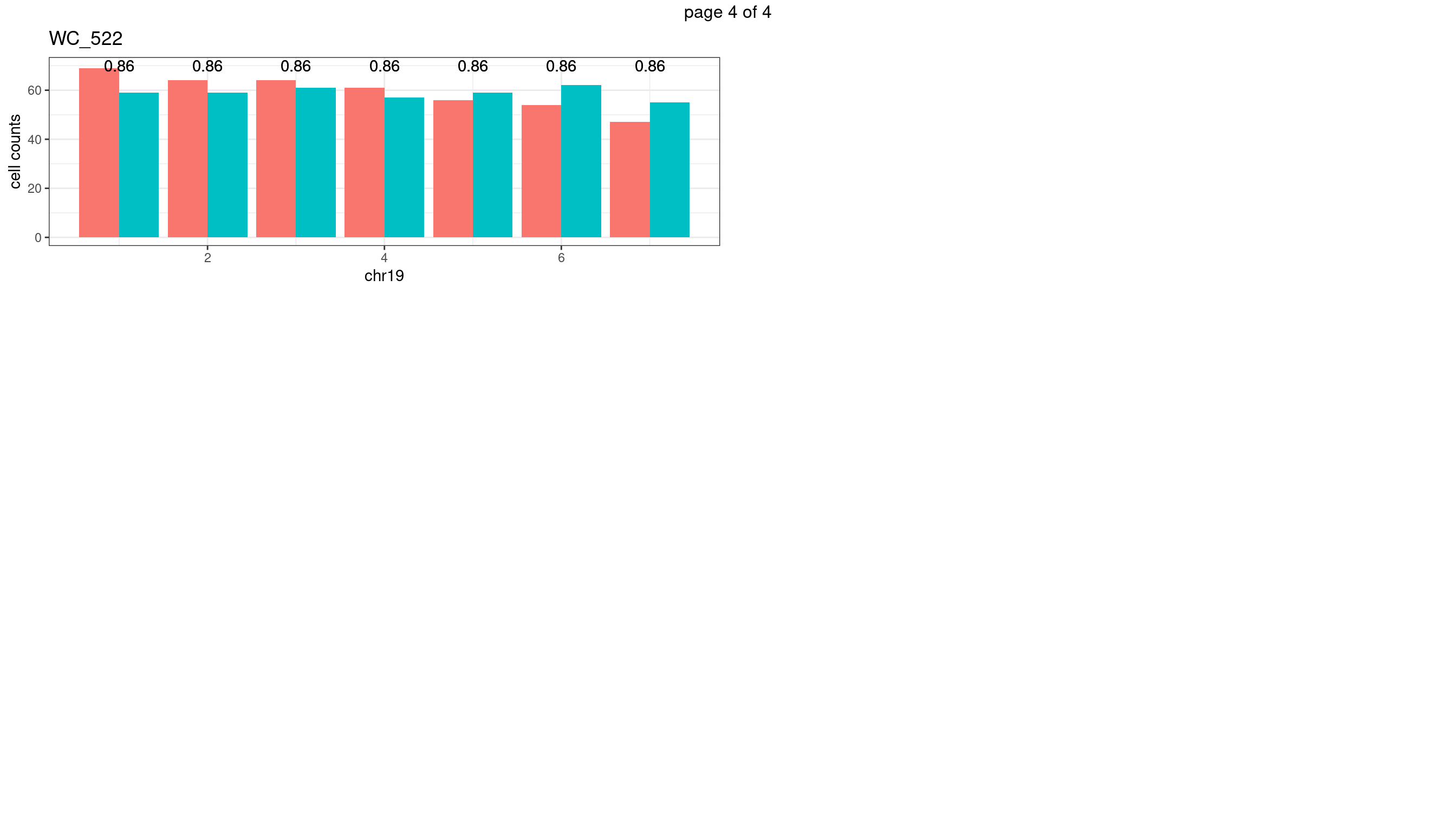
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
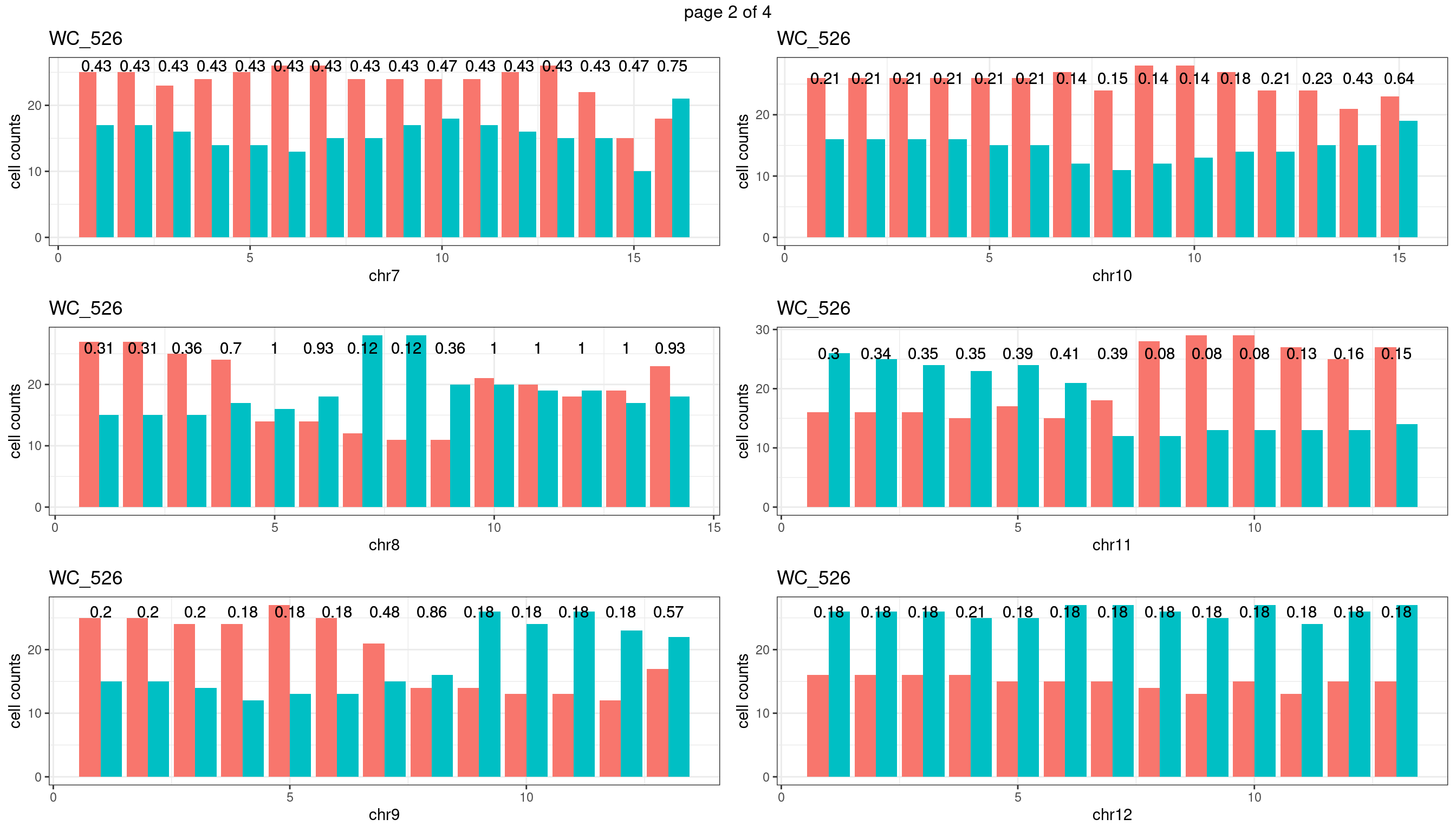
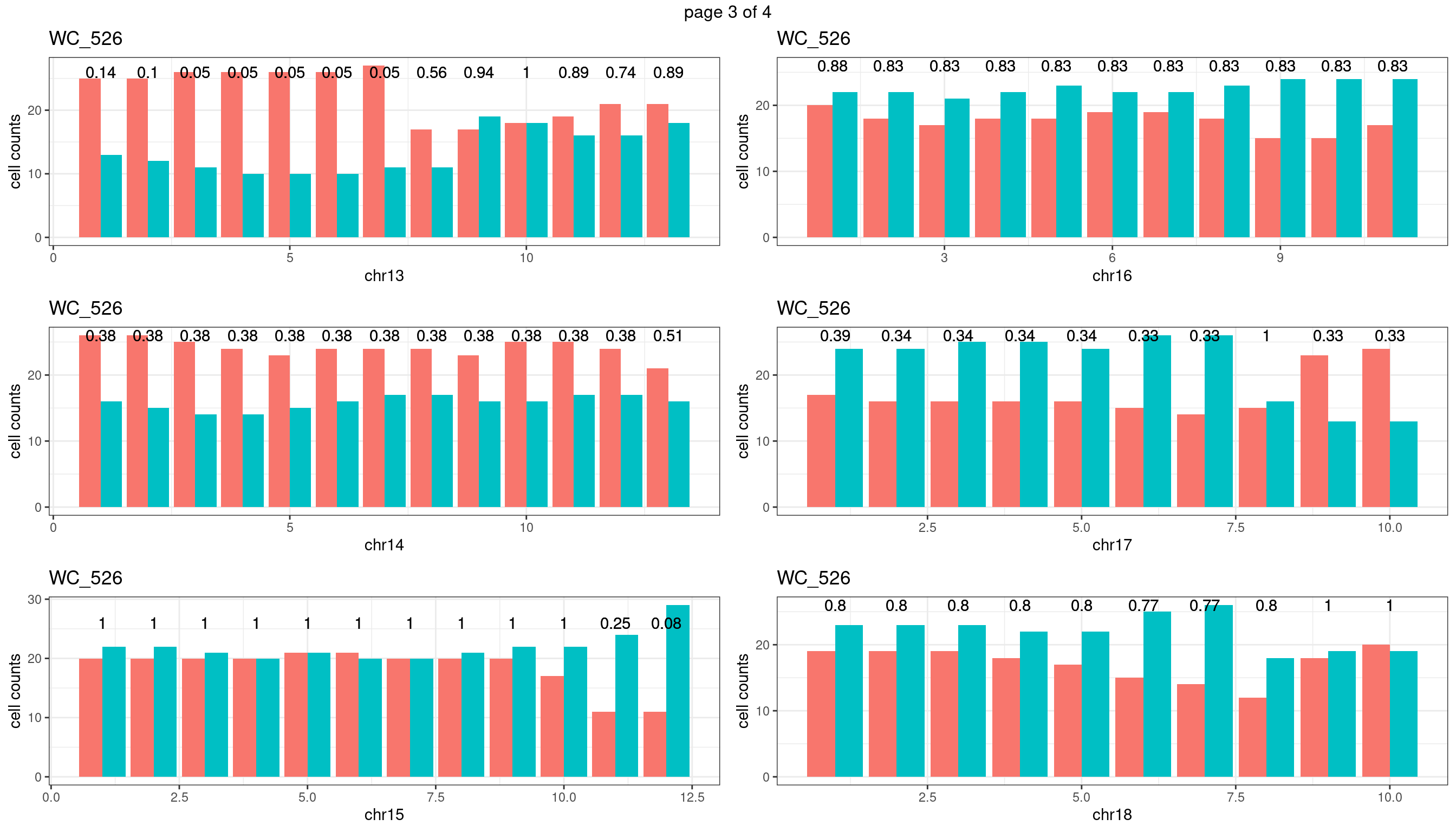
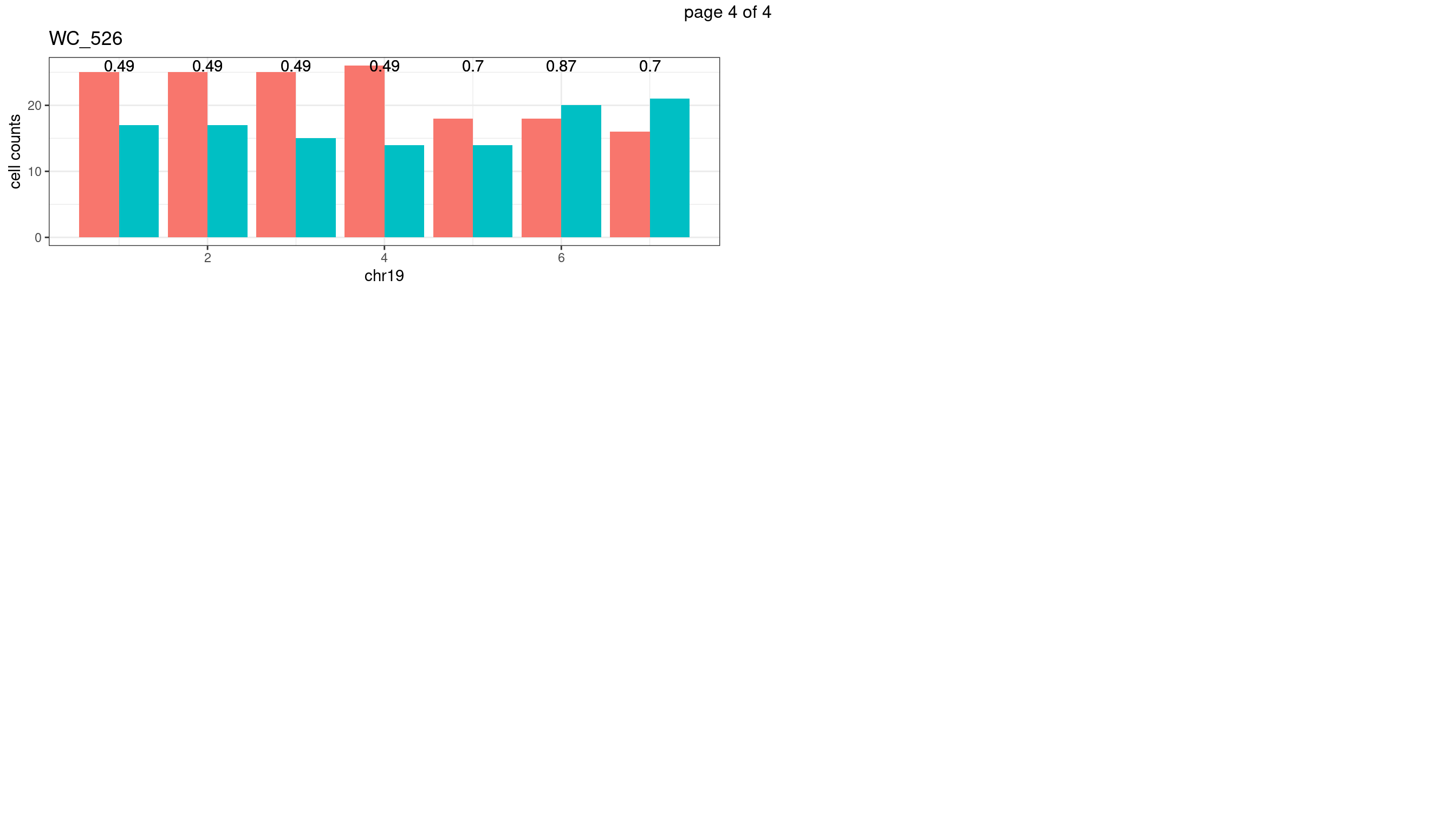
WC_526
sampleName <- "WC_526"
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
stat = "identity")+
geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)
plots_list[[chrName]] <- p+guides(fill = "none" )
} mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
mChrThresPlots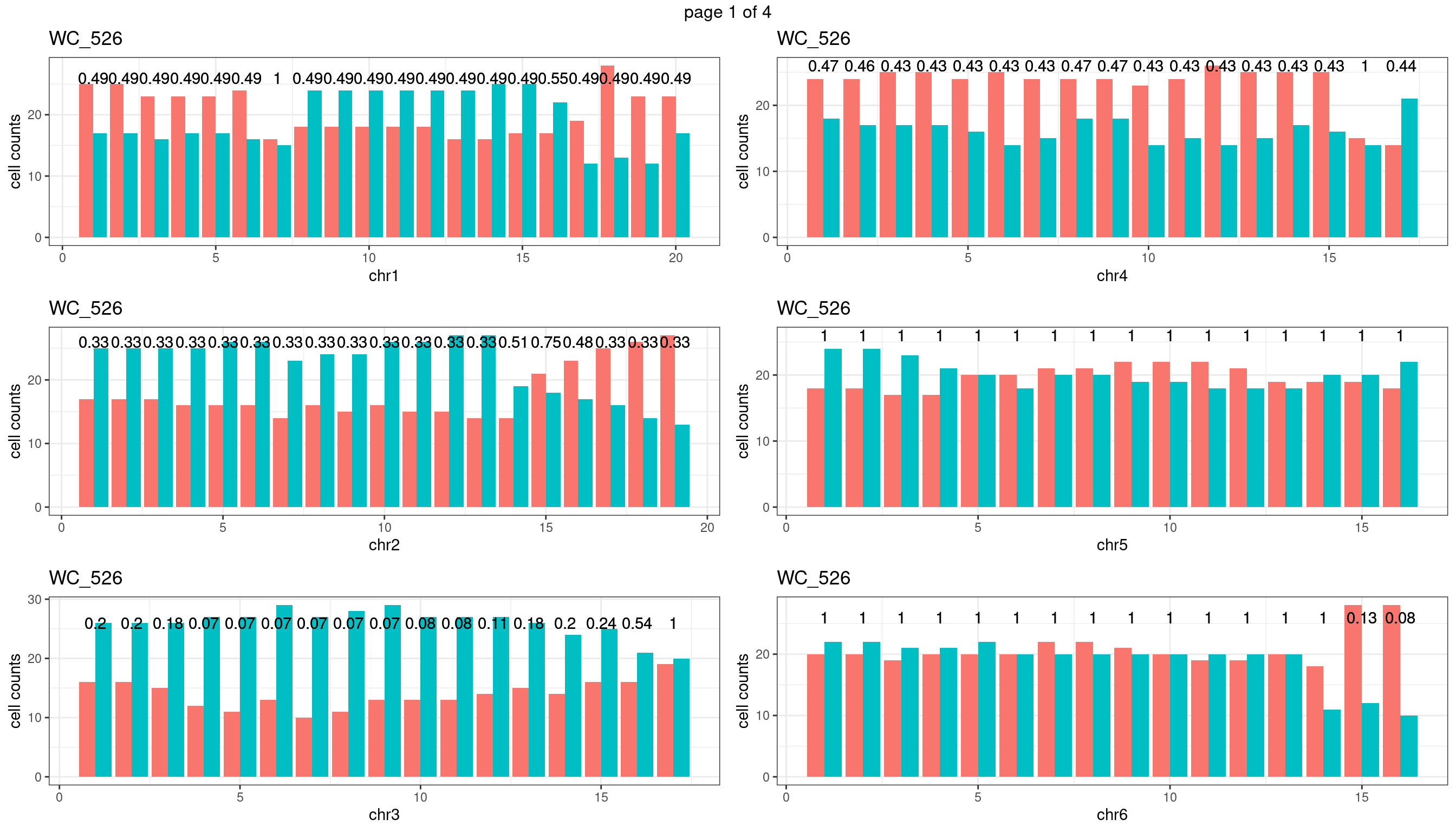
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
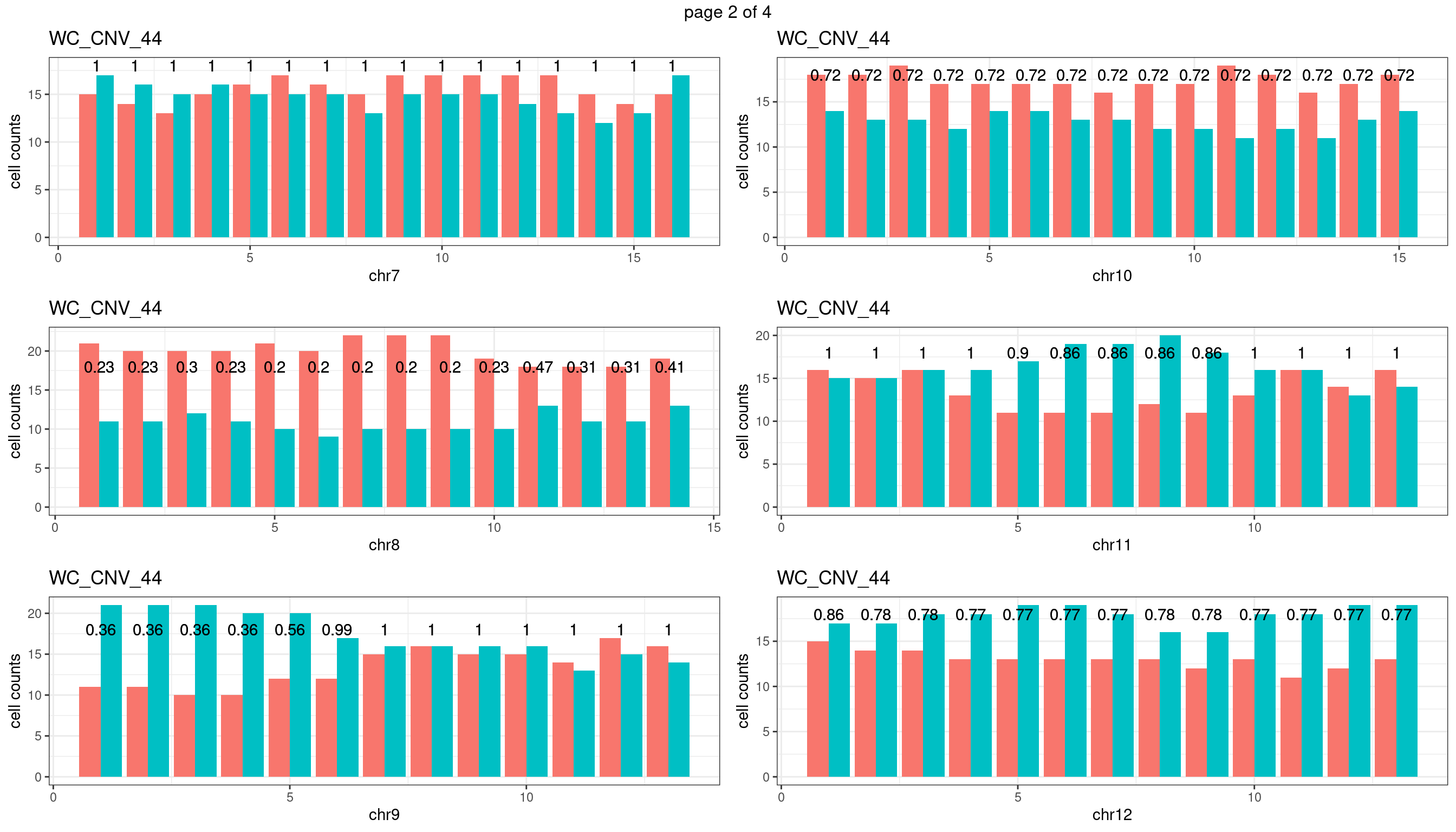
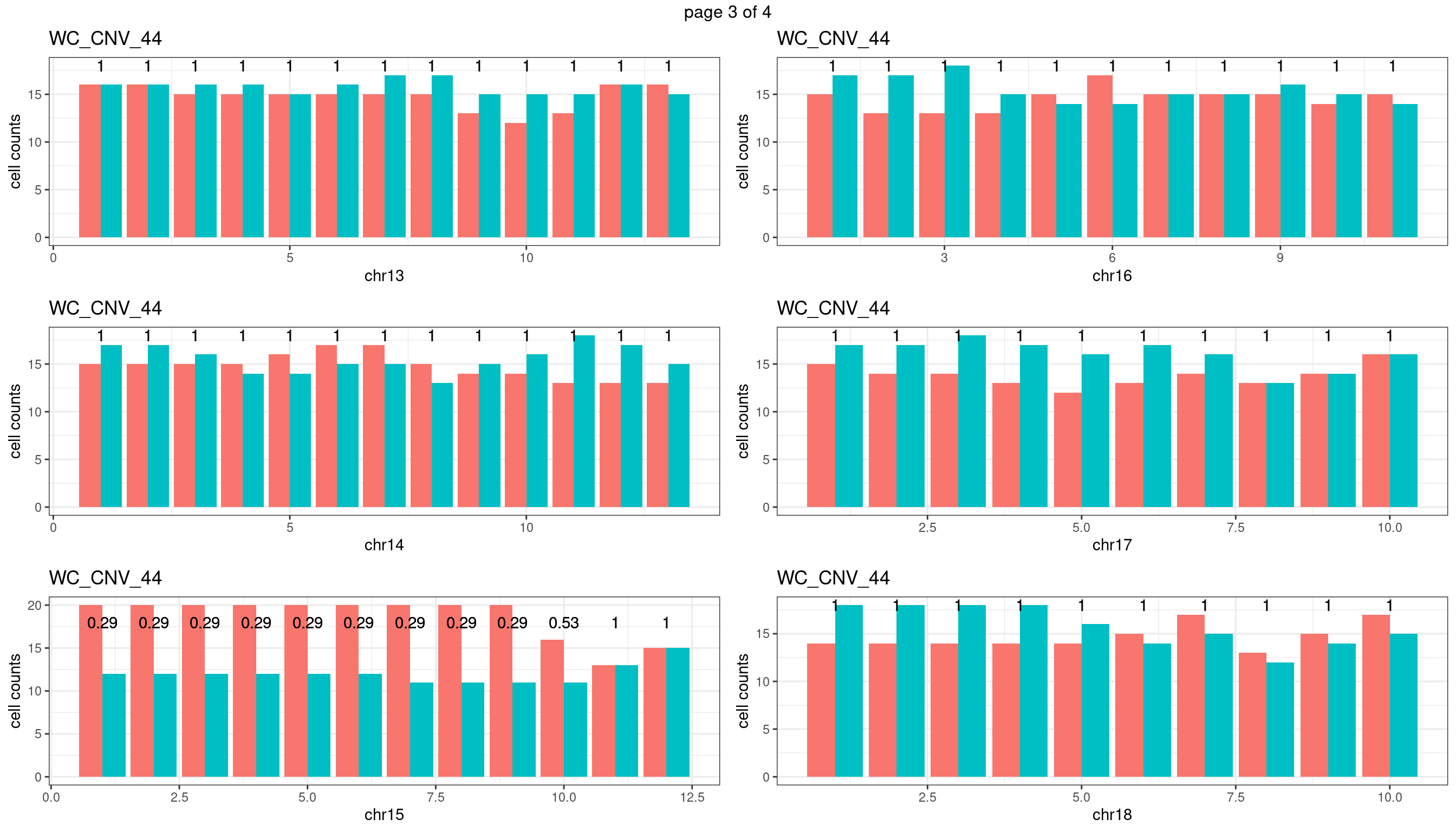
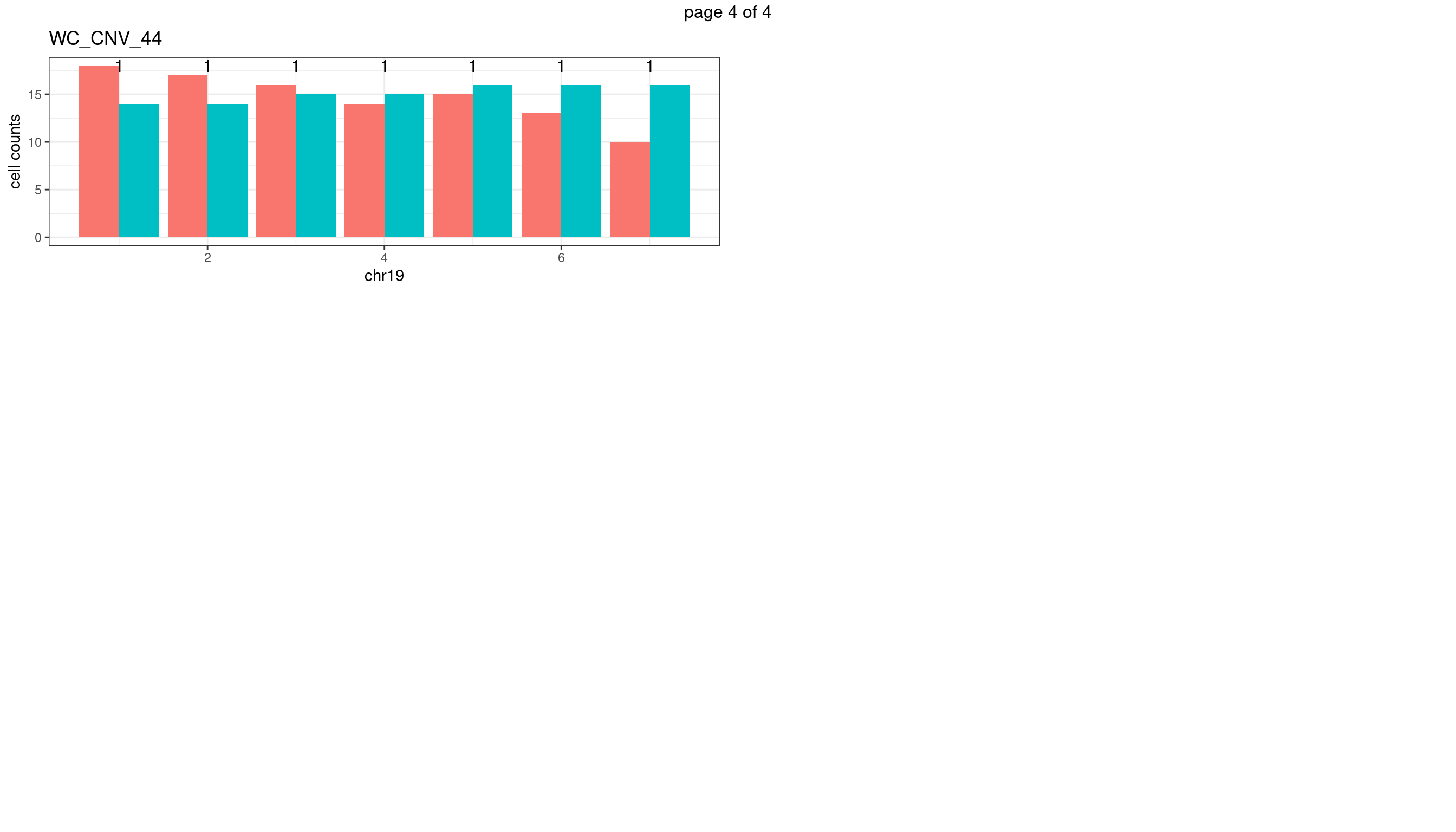
WC_CNV_44
Produce the plots for all chrs
sampleName <- "WC_CNV_44"
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
stat = "identity")+
geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)
plots_list[[chrName]] <- p+guides(fill = "none" )
} mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
mChrThresPlots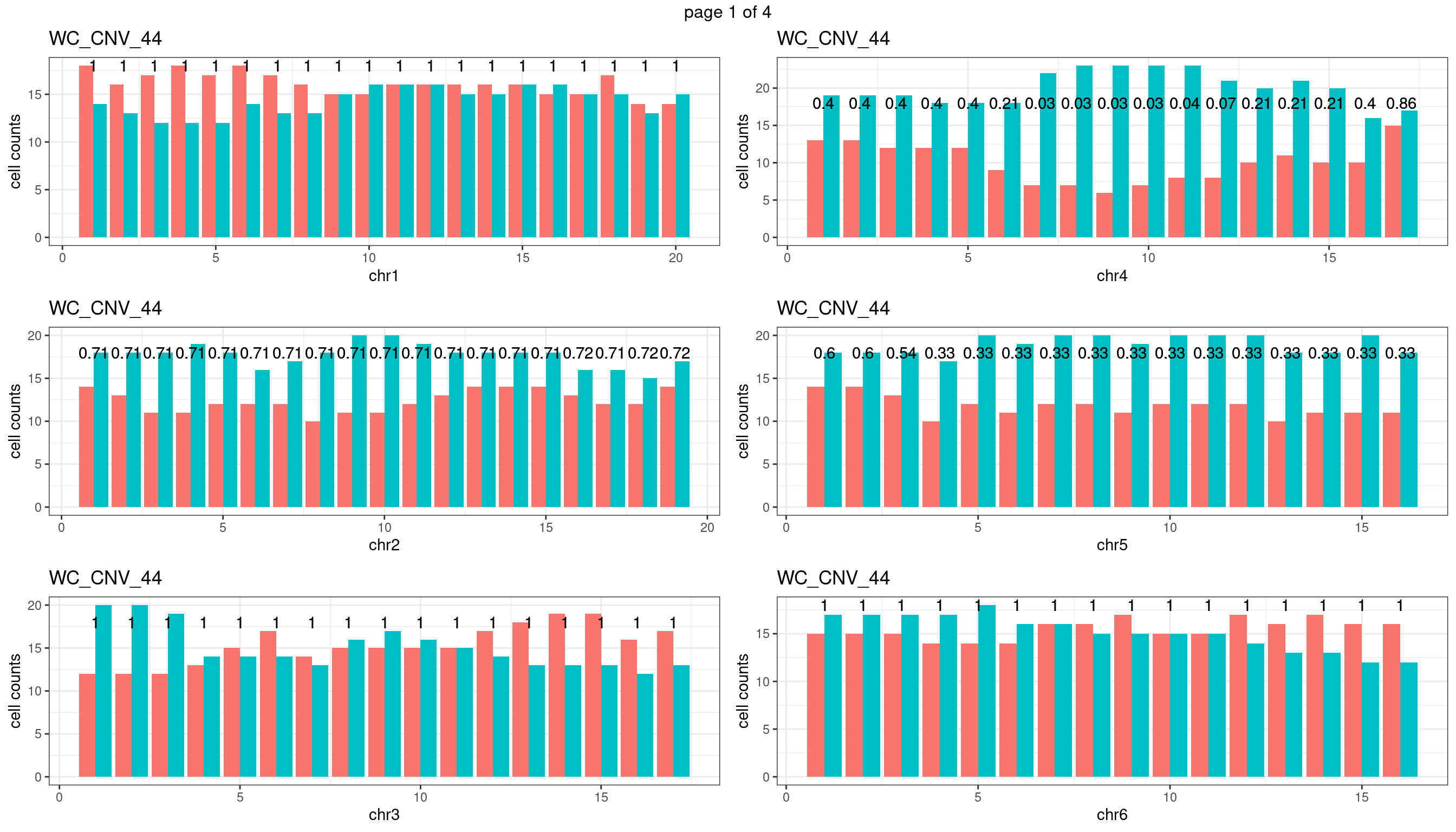
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
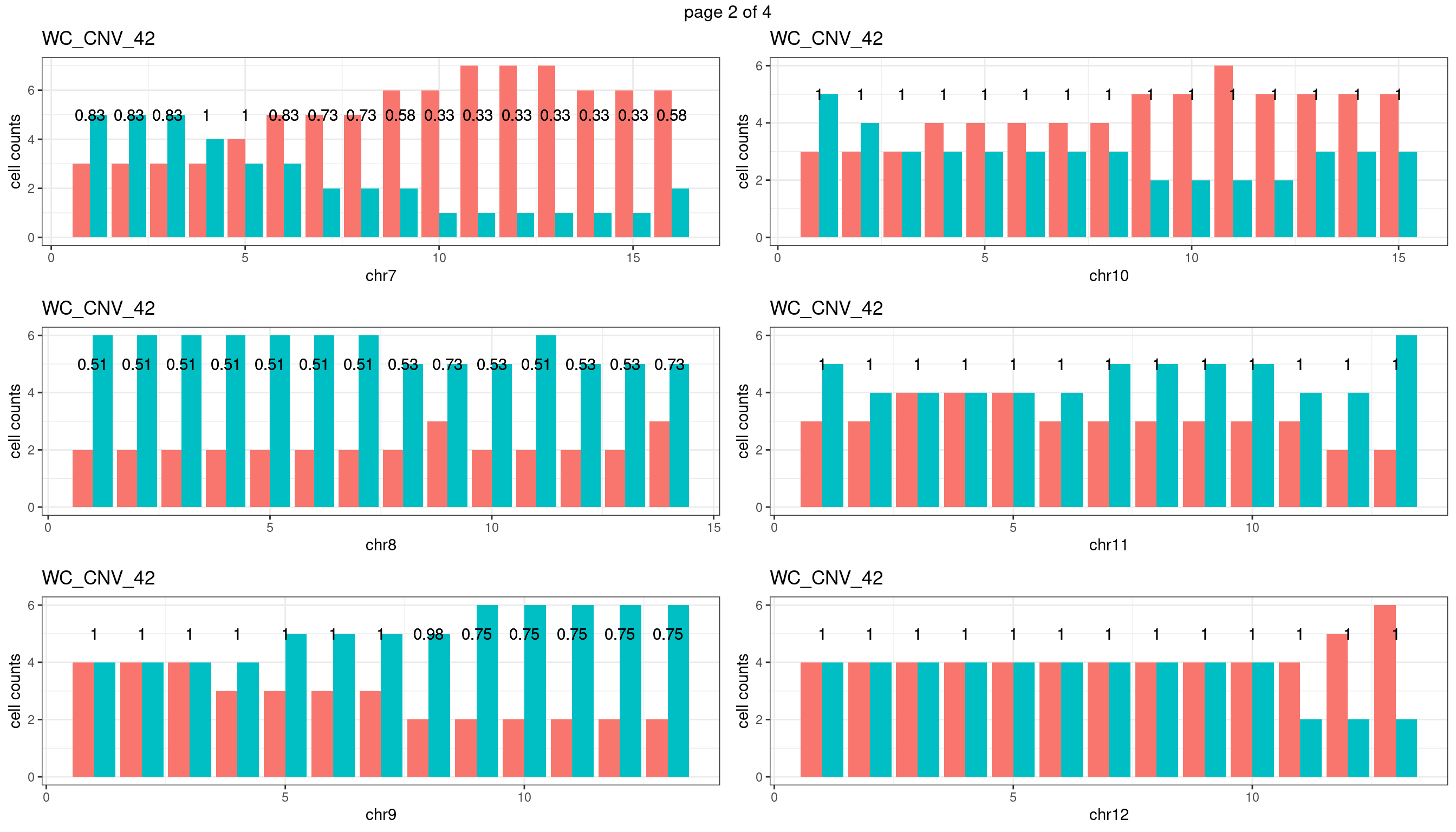
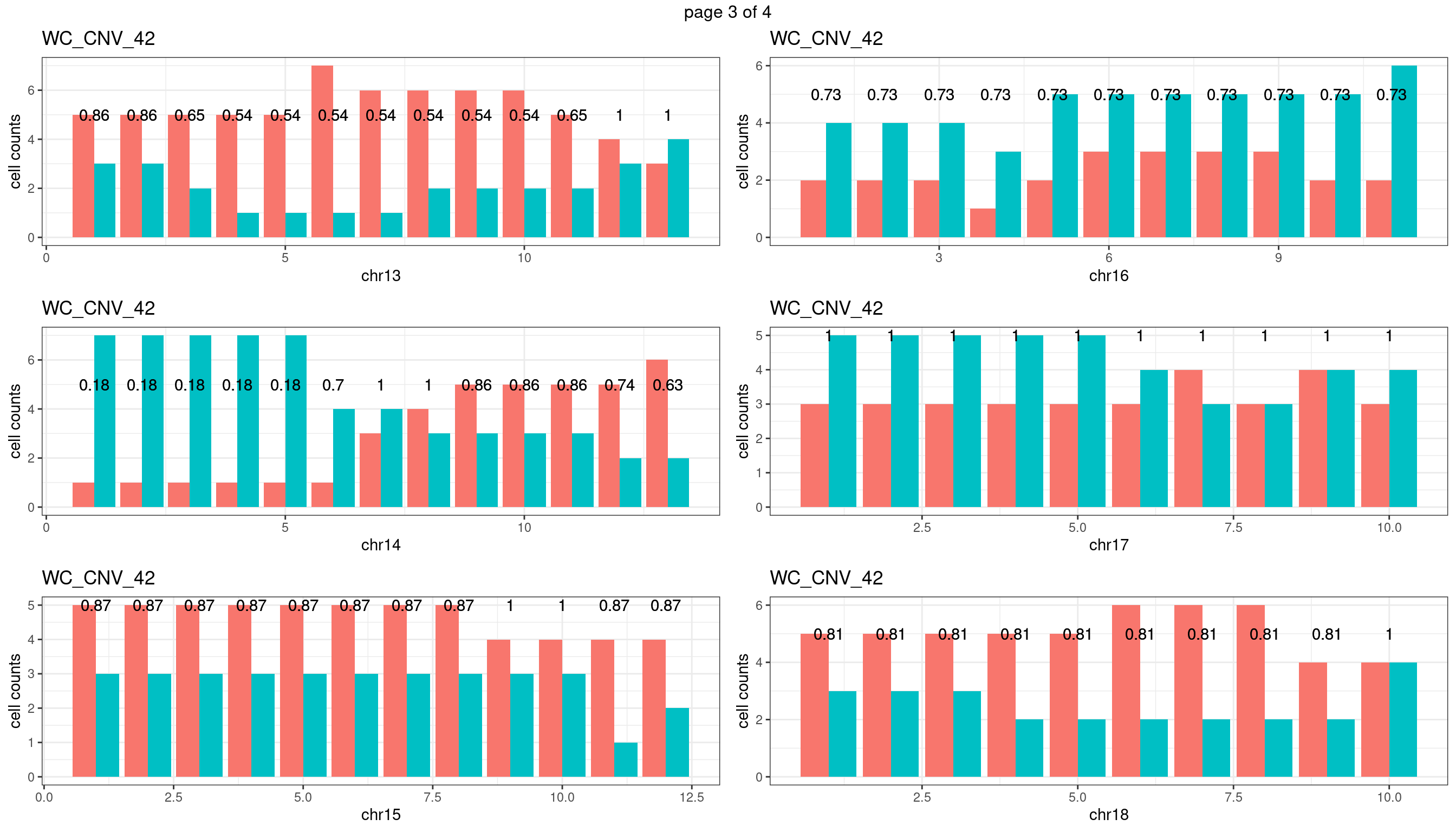
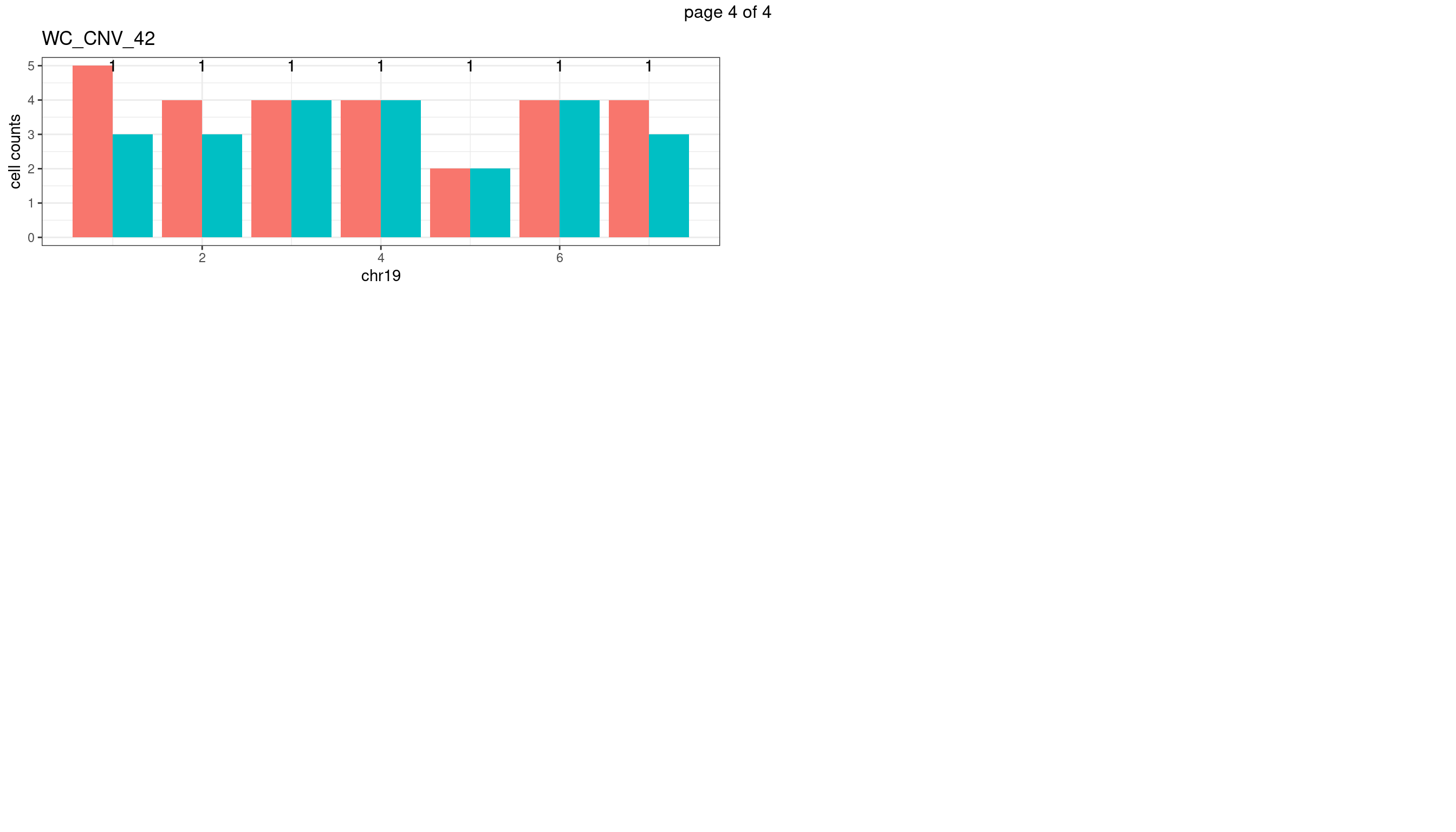
WC_CNV_42
sampleName <- "WC_CNV_42"
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
stat = "identity")+
geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)
plots_list[[chrName]] <- p+guides(fill = "none" )
} mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
mChrThresPlots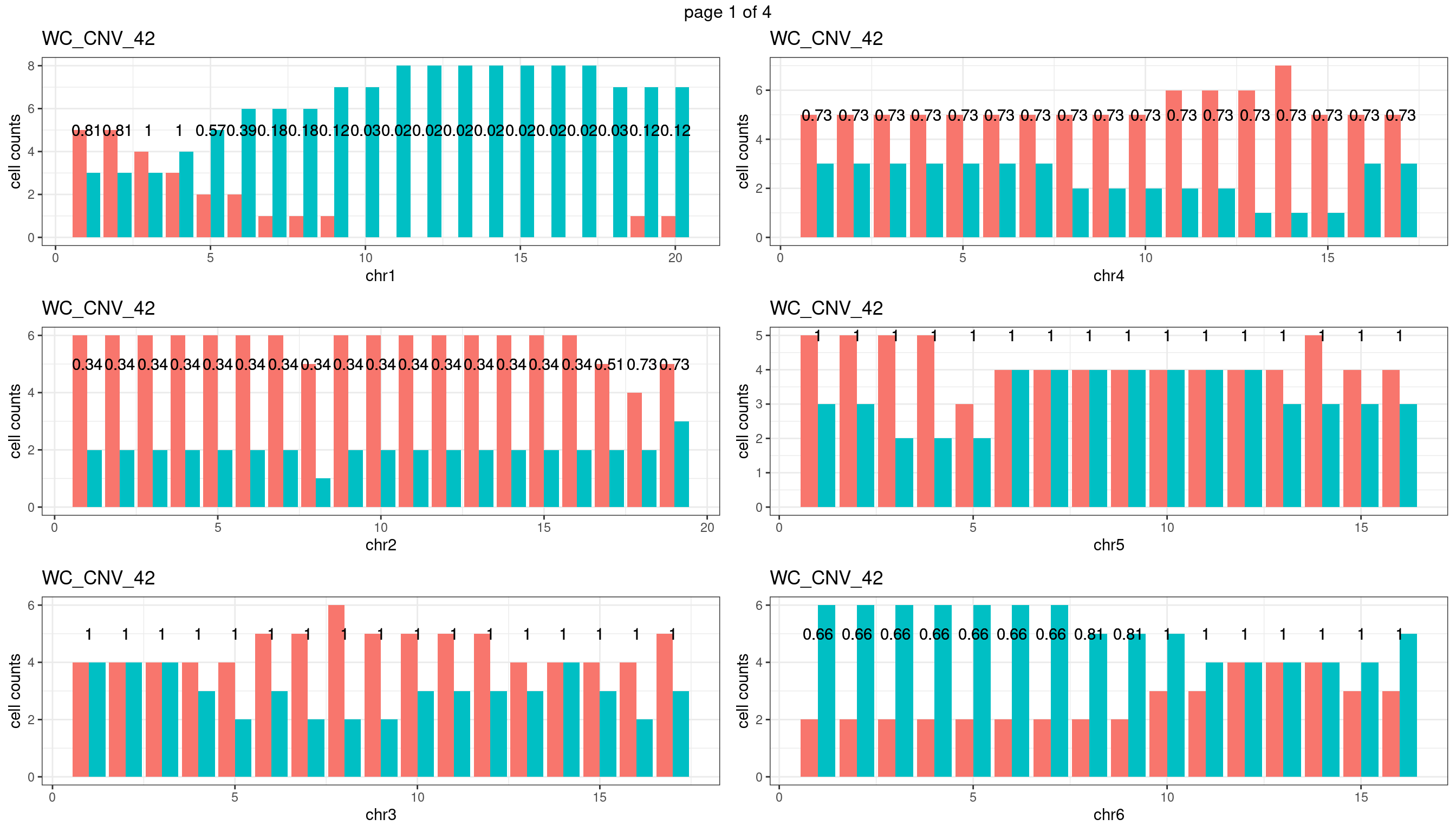
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
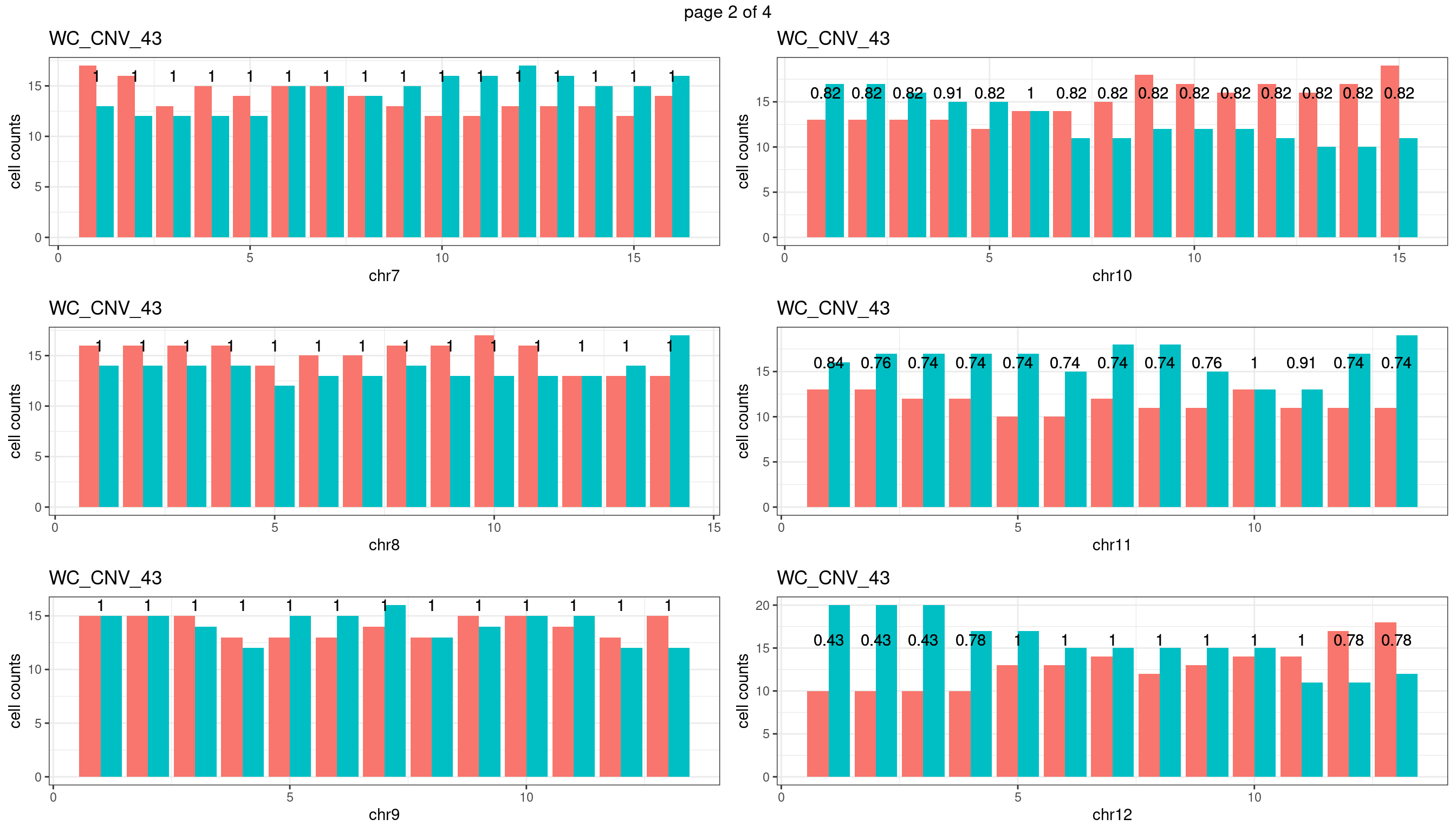
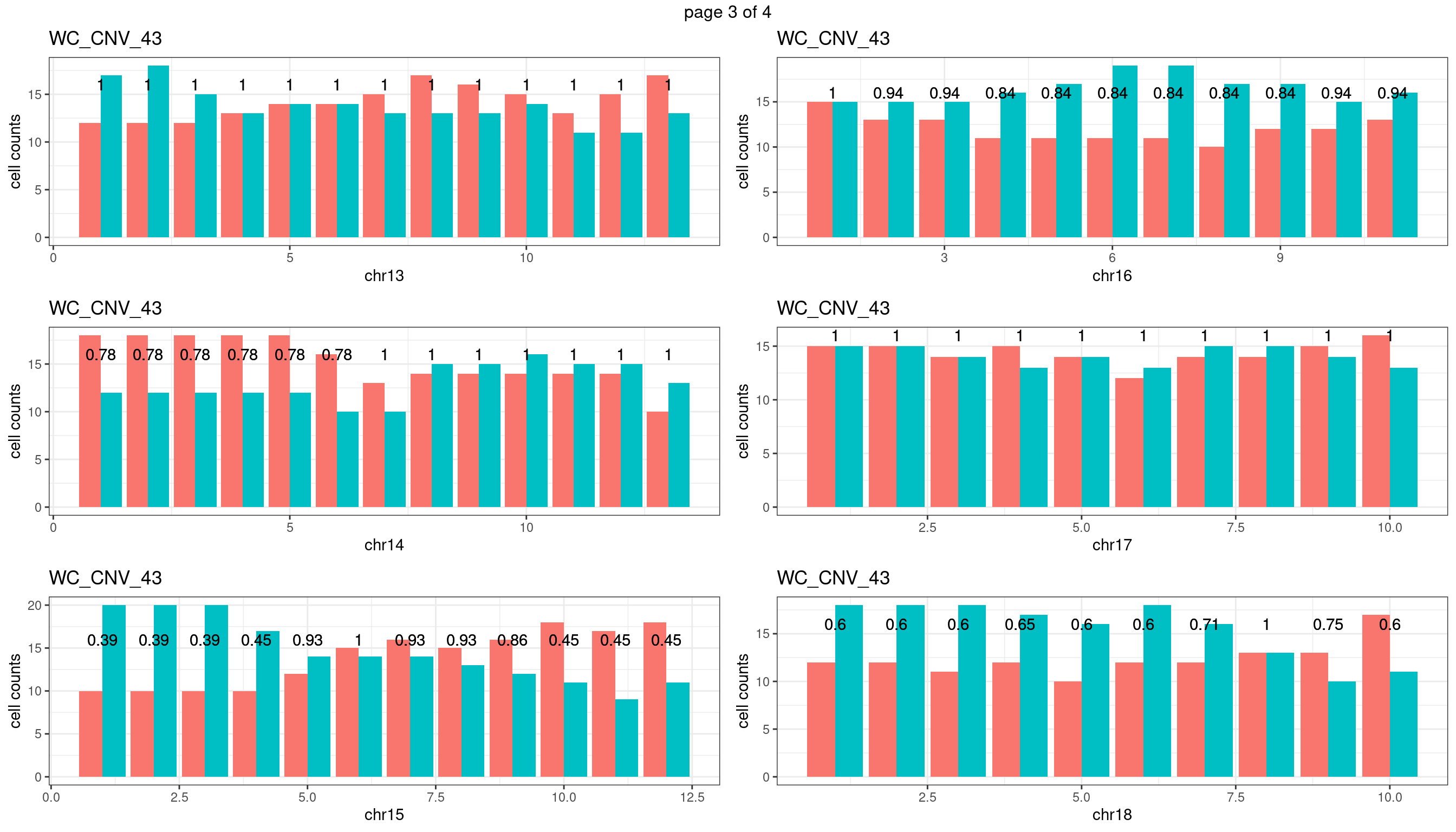
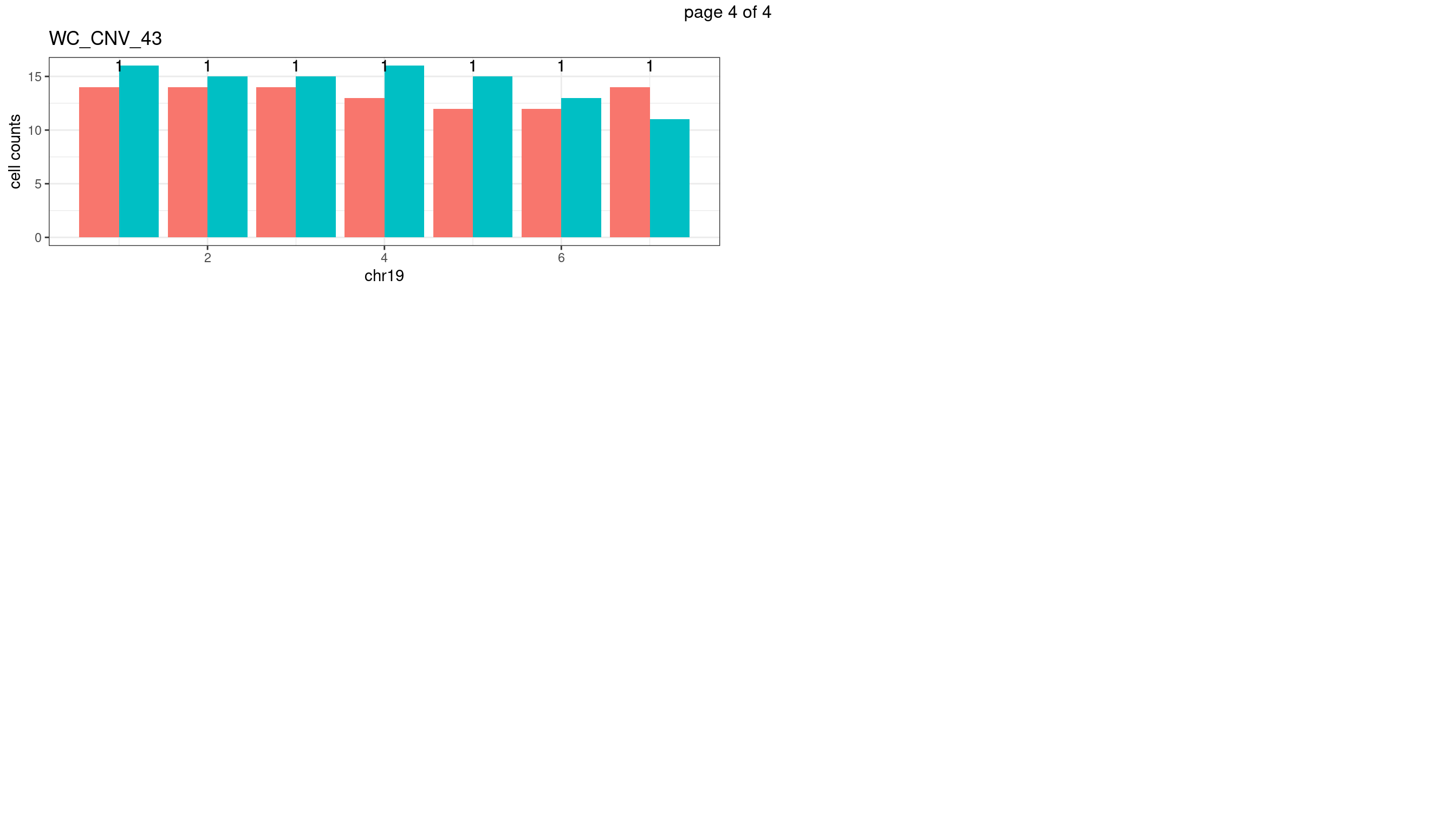
WC_CNV_43
sampleName <- "WC_CNV_43"
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
stat = "identity")+
geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)
plots_list[[chrName]] <- p+guides(fill = "none" )
} mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
mChrThresPlots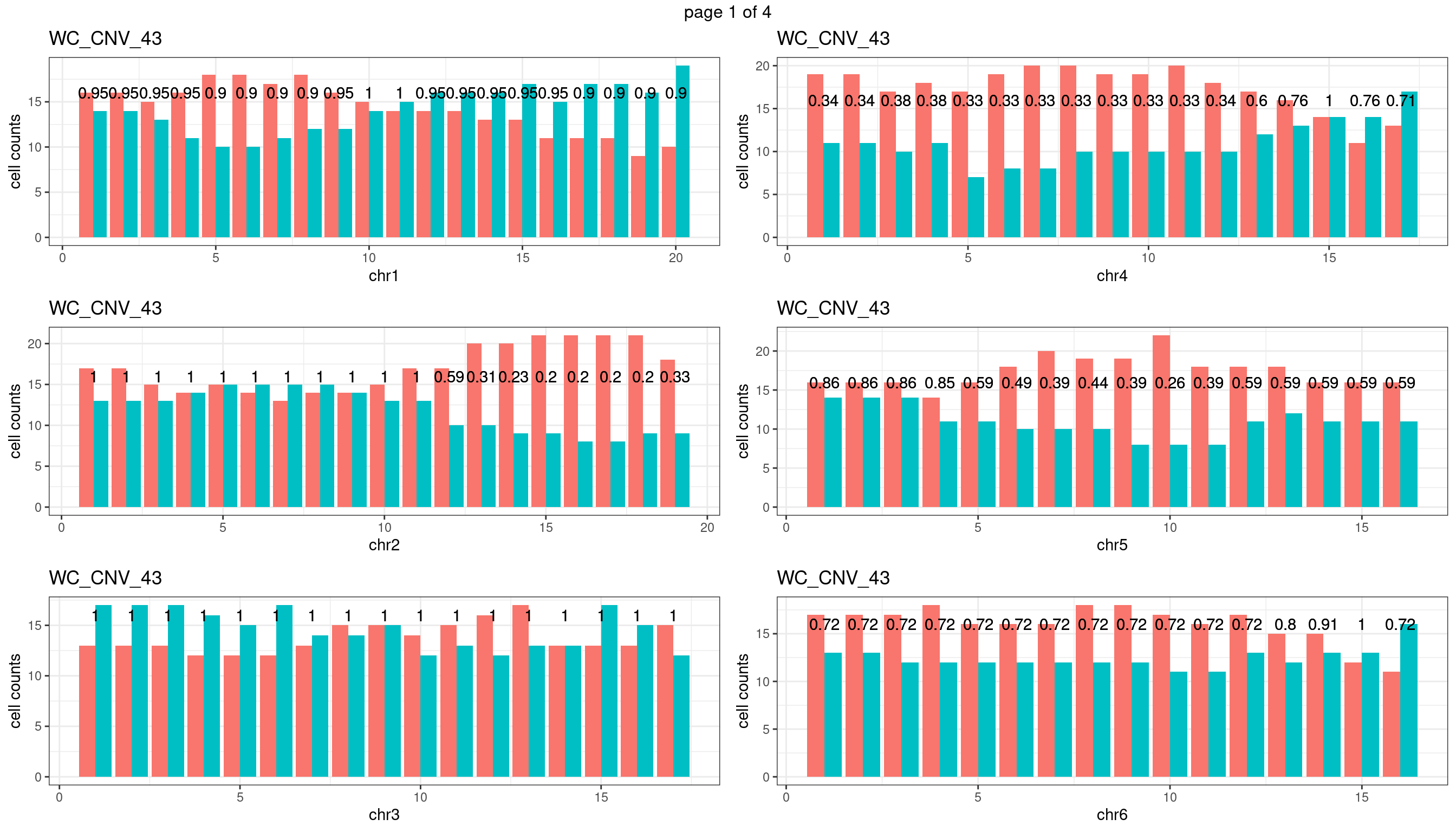
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
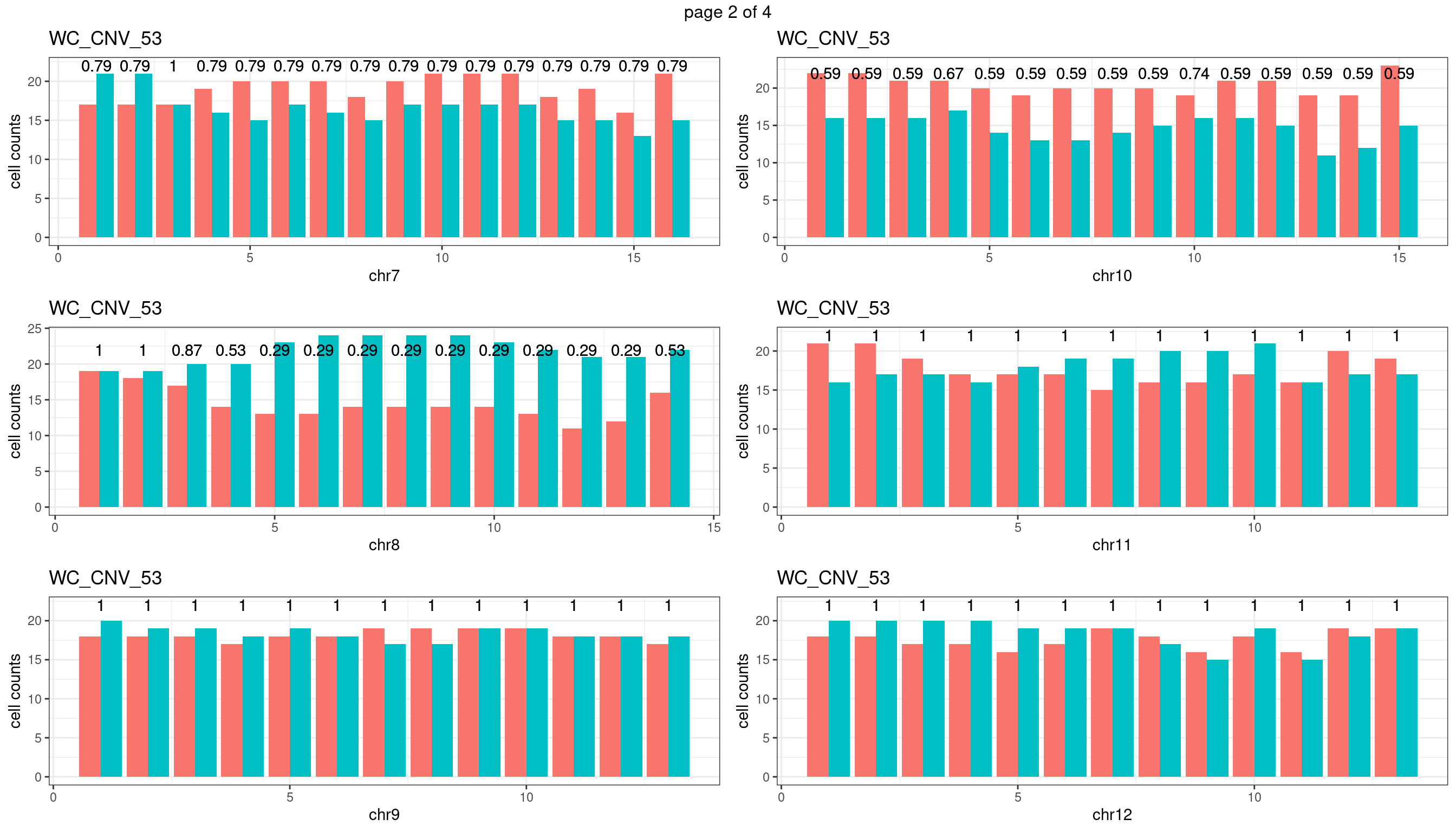
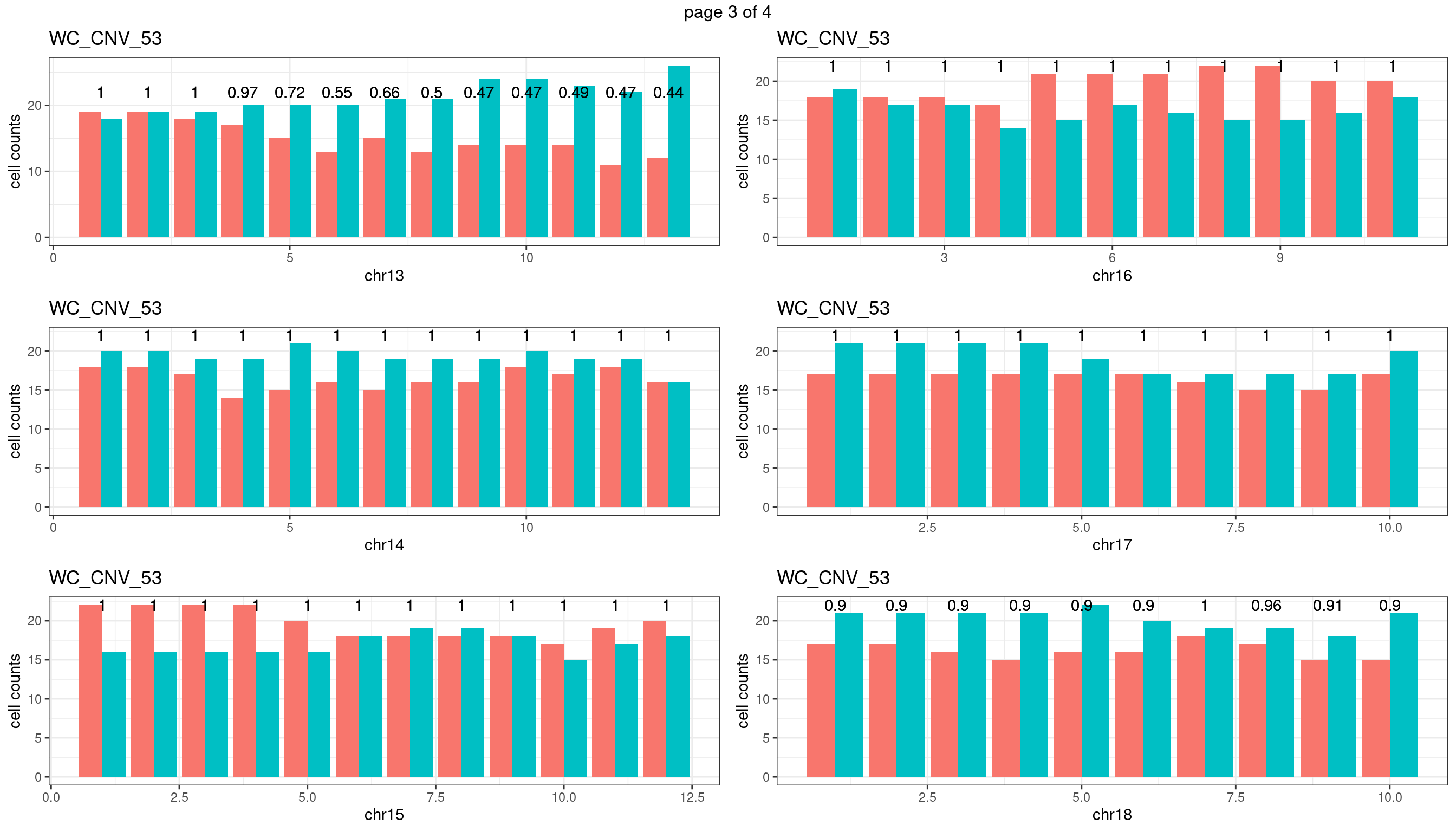
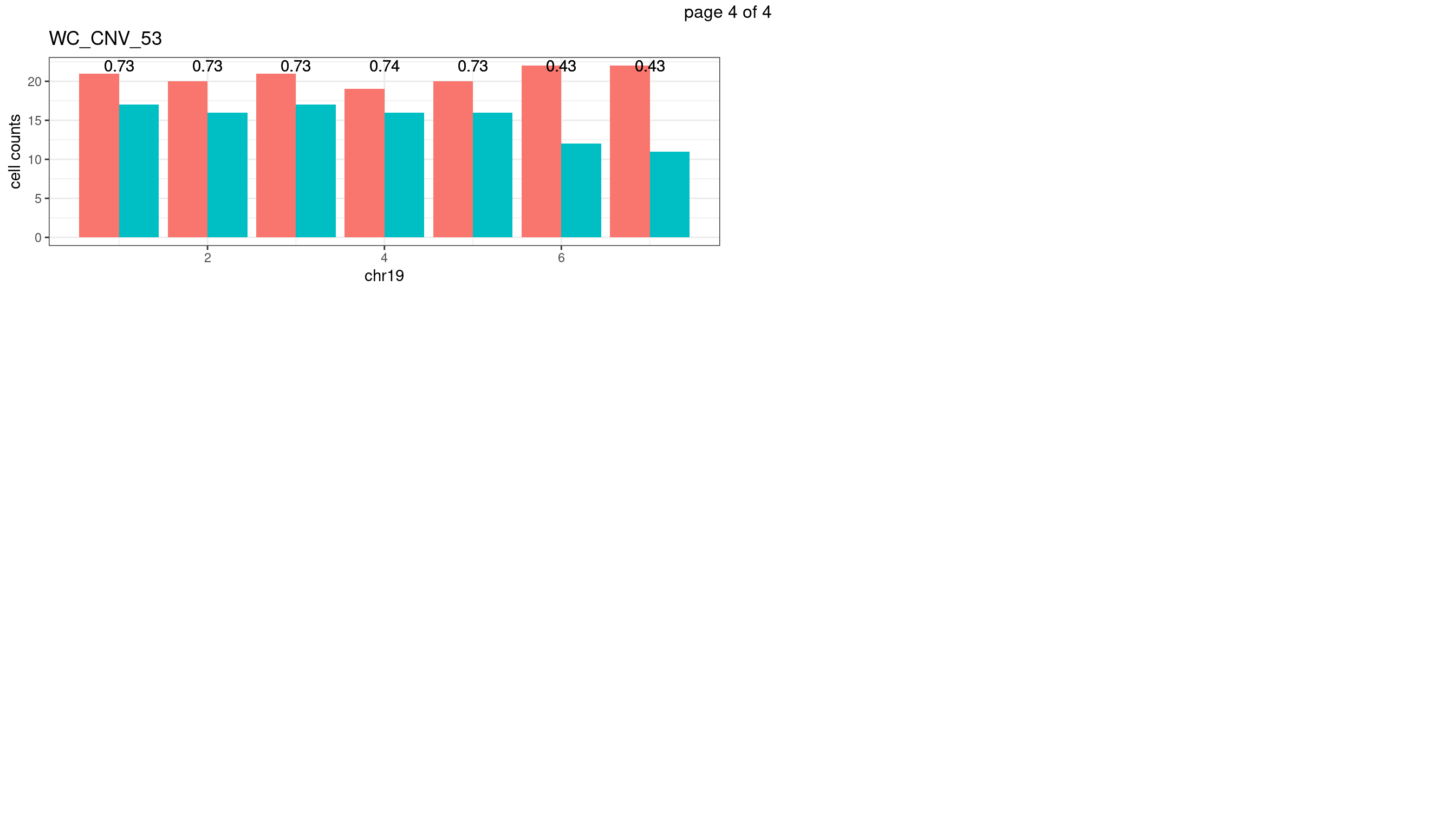
WC_CNV_53
sampleName <- "WC_CNV_53"
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
stat = "identity")+
geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sampleName)
plots_list[[chrName]] <- p+guides(fill = "none" )
} mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
mChrThresPlots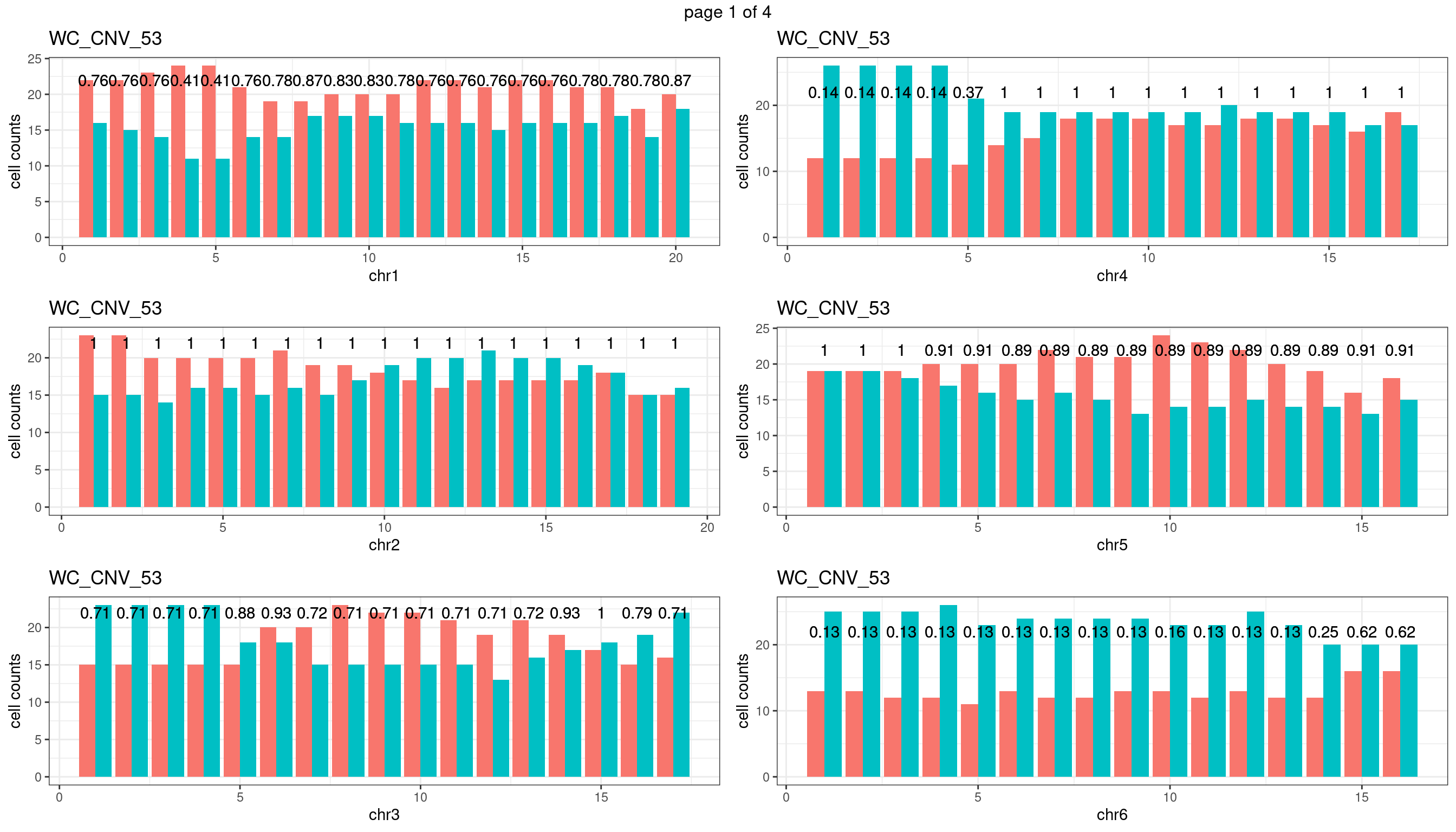
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
Aggregate all cells from mutant mouse individuals
mutant_samples <- c("WC_522","WC_526","WC_CNV_43")#sampleName <- "WC_CNV_44"
plots_list <- list()
bins_pvals_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- lapply(mutant_samples,function(sampleName){
readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
})
merged_mcols <- do.call(cbind,lapply(bin_state_gr, mcols))
s1_counts <- rowSums(data.frame(merged_mcols,check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(merged_mcols,check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
# p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
# geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
# stat = "identity")+
# geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
# xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle("mutant")
p <- plot_df %>% mutate(hap_ratio = s1_count /(s1_count +s2_count )) %>% ggplot() +
geom_point(aes(x = bin_id, y = hap_ratio))+geom_hline(mapping = aes(yintercept = 0.5),linetype="dotted")+
# geom_text(aes(x = bin_id,y=max(hap_ratio)+0.1, label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+ylim(c(0,1))+theme_bw(base_size = 18)+ggtitle("mutant")
plots_list[[chrName]] <- p+guides(fill = "none" )
bins_pvals_list[[chrName]] <- bins_pvals
}
message(mutant_samples, " number of bins with FDR < 0.05: ",
sum(p.adjust(unlist(bins_pvals_list),"fdr")<0.05))WC_522WC_526WC_CNV_43 number of bins with FDR < 0.05: 0notitle_p <- lapply(plots_list, function(x) x +ggtitle("")+ylab("")+theme(plot.margin = unit(c(0.00,0.00,-0.02,0.00), "cm")))
mChrThresPlots <- marrangeGrob(notitle_p, ncol=7,nrow=3,
left = textGrob("Haplotype state ratio",
rot = 90,gp = gpar(fontsize=22)),
layout_matrix = matrix(c(1:19,NA,NA), 3, 7, TRUE),
right= " ")
mChrThresPlots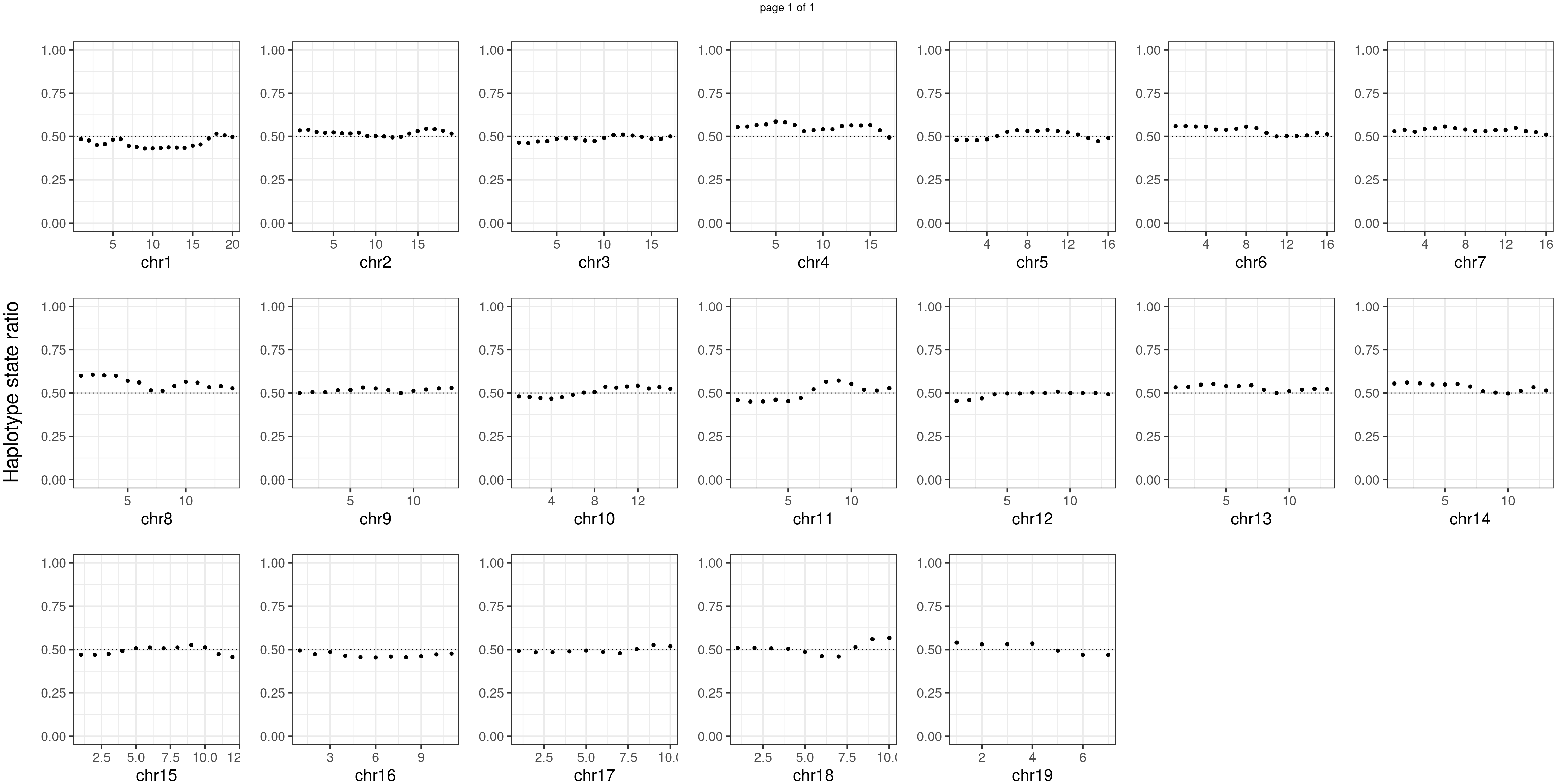
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
Aggregate all cells from wildtype mouse individuals
wildtype_samples <- c("WC_CNV_42","WC_CNV_44","WC_CNV_53")
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- lapply(wildtype_samples,function(sampleName){
readRDS(paste0("./output/outputR/analysisRDS/",sampleName,"_",
chrName,"bin_state_gr.rds"))
})
merged_mcols <- do.call(cbind,lapply(bin_state_gr, mcols))
s1_counts <- rowSums(data.frame(merged_mcols,check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(merged_mcols,check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
# p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
# geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
# stat = "identity")+
# geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
# xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle("mutant")
p <- plot_df %>% mutate(hap_ratio = s1_count /(s1_count +s2_count )) %>% ggplot() +
geom_point(aes(x = bin_id, y = hap_ratio))+geom_hline(mapping = aes(yintercept = 0.5),linetype="dotted")+
# geom_text(aes(x = bin_id,y=max(hap_ratio)+0.1, label = round(fdr,2)))+
xlab(chrName)+ylab("cell counts")+ylim(c(0,1))+theme_bw(base_size = 18)+ggtitle("mutant")
plots_list[[chrName]] <- p+guides(fill = "none" )
} notitle_p <- lapply(plots_list, function(x) x +ggtitle("")+ylab("")+theme(plot.margin = unit(c(0.00,0.00,-0.02,0.00), "cm")))
mChrThresPlots <- marrangeGrob(notitle_p, ncol=7,nrow=3,
left = textGrob("Haplotype state ratio",
rot = 90,gp = gpar(fontsize=22)),
layout_matrix = matrix(c(1:19,NA,NA), 3, 7, TRUE),
right= " ")
mChrThresPlots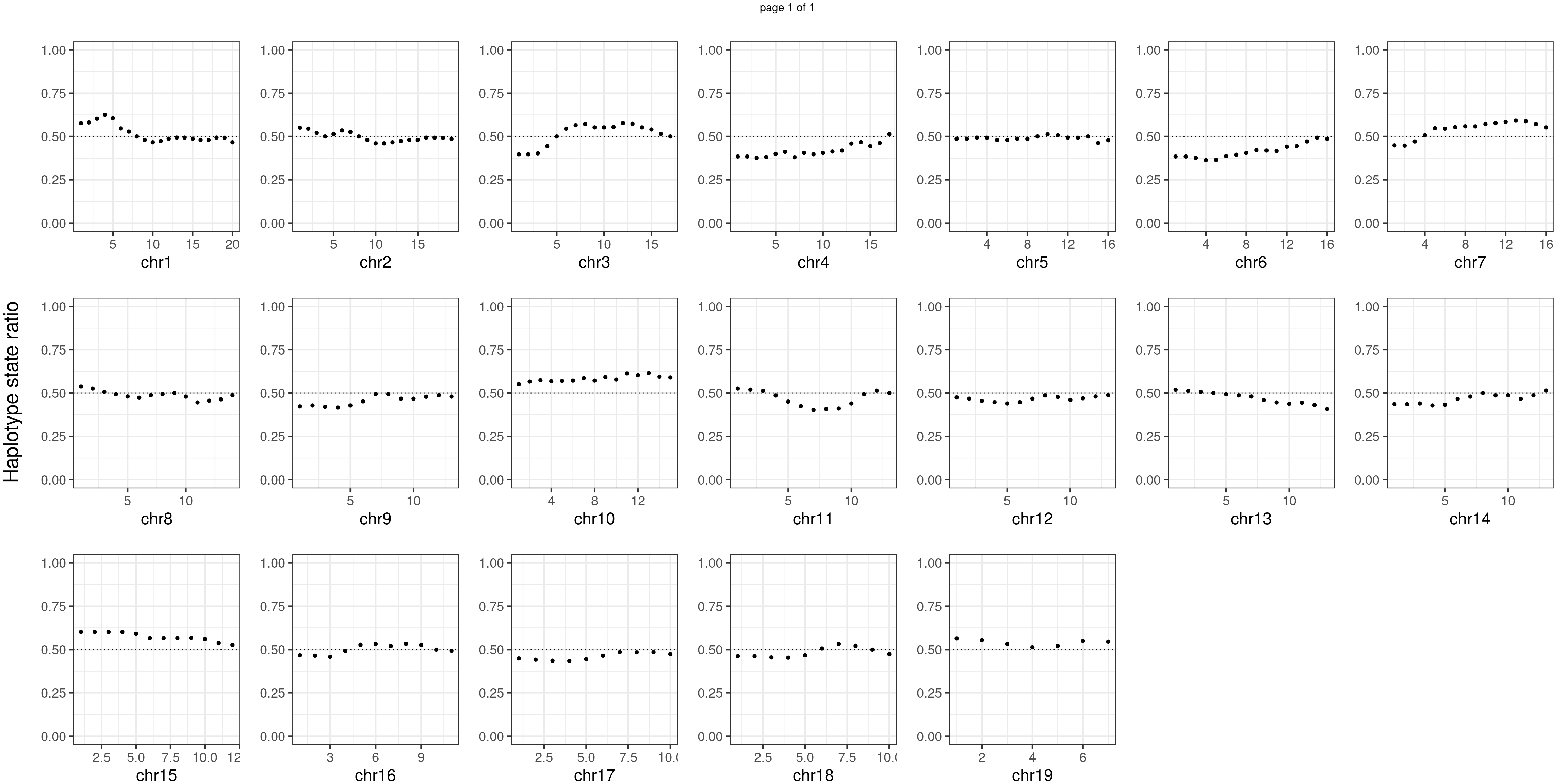
| Version | Author | Date |
|---|---|---|
| 9a3ae70 | rlyu | 2021-08-05 |
Conclusion
There is no apparent regions with imbalanced segregation among the sperm cells from mutant and sperm cells from wildtype.
BC1F1 samples
Similar idea for BC1F1 samples.
plots_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/bc1f1_",
chrName,"bin_state_gr.rds"))
s1_counts <- rowSums(data.frame(bin_state_gr,check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(bin_state_gr,check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
# p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
# geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
# stat = "identity")+
# geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
# xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle("all bc1f1s")
p <- plot_df %>% mutate(hap_ratio = s1_count /(s1_count +s2_count )) %>% ggplot() +
geom_point(aes(x = bin_id, y = hap_ratio))+geom_hline(mapping = aes(yintercept = 0.5),linetype="dotted")+
# geom_text(aes(x = bin_id,y=max(hap_ratio)+0.1, label = round(fdr,2)))+
xlab(chrName)+ylab("sample counts")+ylim(c(0,1))+theme_bw(base_size = 18)+ggtitle("all bc1f1s")
plots_list[[chrName]] <- p+guides(fill = "none" )
} notitle_p <- lapply(plots_list, function(x) x +ggtitle("")+ylab("")+
theme(plot.margin = unit(c(0.00,0.00,-0.02,0.00), "cm")))
mChrThresPlots <- marrangeGrob(notitle_p, ncol=7,nrow=3,
left = textGrob("Haplotype state ratio",
rot = 90,gp = gpar(fontsize=22)),
layout_matrix = matrix(c(1:19,NA,NA), 3, 7, TRUE),
right= " ")
mChrThresPlots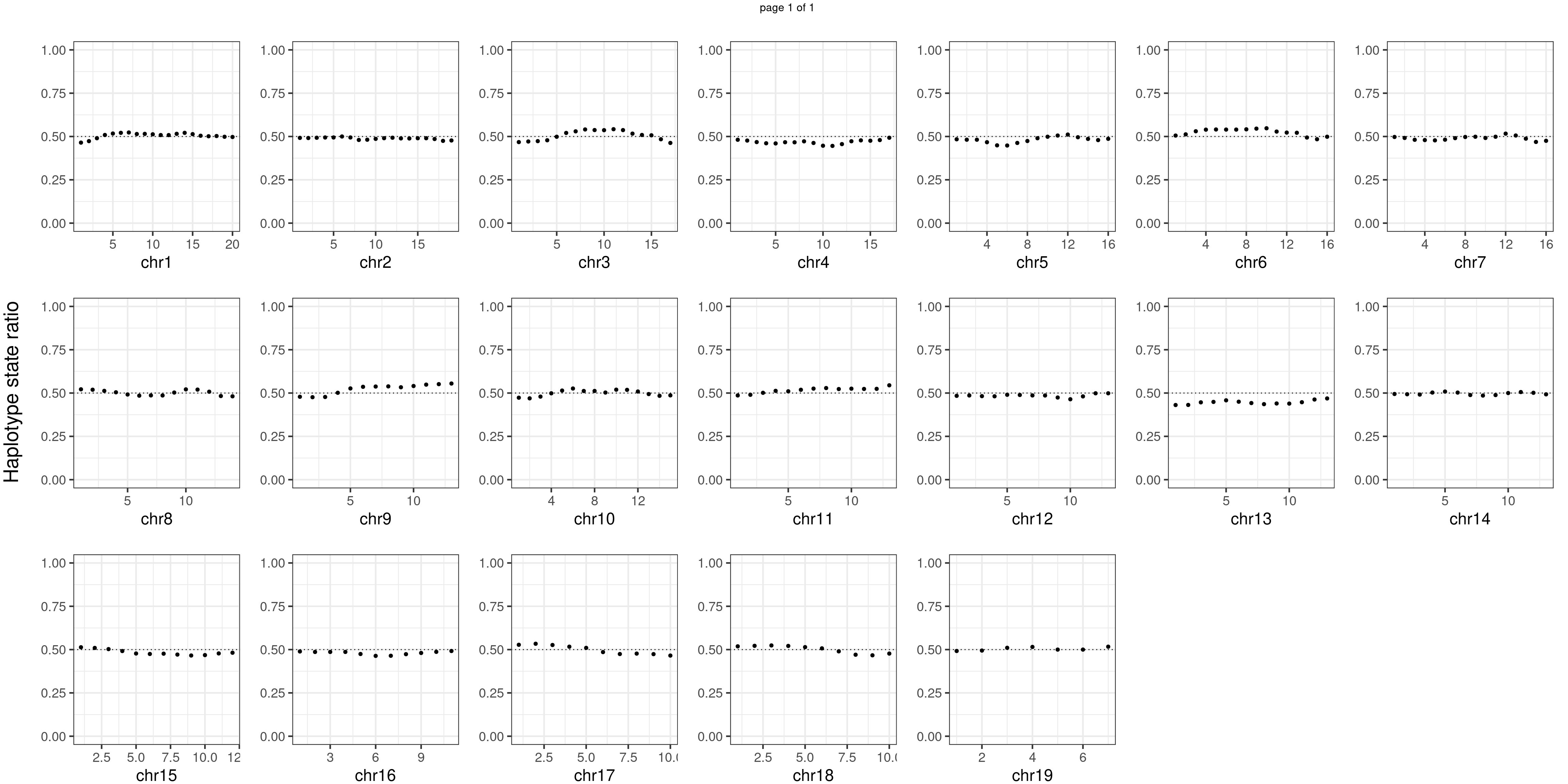
Per BC1F1 sample group
co_count <- readRDS(file = "output/outputR/analysisRDS/all_rse_count_07-20.rds")for(sample_group in unique(co_count$sampleGroup) ){
plots_list <- list()
bins_pvals_list <- list()
for(chrName in paste0("chr",1:19)){
bin_state_gr <- readRDS(paste0("./output/outputR/analysisRDS/bc1f1_",
chrName,"bin_state_gr.rds"))
group_sids <- colnames(mcols(bin_state_gr)) %in% co_count$Sid[co_count$sampleGroup==sample_group]
mcols(bin_state_gr) <- mcols(bin_state_gr)[group_sids]
s1_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s1",na.rm=T)
s2_counts <- rowSums(data.frame(mcols(bin_state_gr),check.names = F)=="s2",na.rm=T)
bins_pvals <- lapply(seq(s1_counts), function(i){
btest <- binom.test(c(s1_counts[i],s2_counts[i]))
btest$p.value
})
plot_df <- data.frame(s1_count = s1_counts,
s2_count = s2_counts,
bin_id = seq(length(bins_pvals)),
biom_pvals = unlist(bins_pvals),
fdr = p.adjust(unlist(bins_pvals),method = "fdr"))
# p <- plot_df %>% tidyr::pivot_longer(c("s1_count","s2_count")) %>% ggplot() +
# geom_bar(aes(x = bin_id, y = value,fill = name),position = "dodge",
# stat = "identity")+
# geom_text(aes(x = bin_id,y=max(c(s1_counts,s2_counts)), label = round(fdr,2)))+
# xlab(chrName)+ylab("cell counts")+theme_bw()+ggtitle(sample_group)
p <- plot_df %>% mutate(hap_ratio = s1_count /(s1_count +s2_count )) %>% ggplot() +
geom_point(aes(x = bin_id, y = hap_ratio))+geom_hline(mapping = aes(yintercept = 0.5),linetype="dotted")+
geom_text(aes(x = bin_id,y=max(hap_ratio)+0.1, label = round(fdr,2)))+
xlab(chrName)+ylab("sample counts")+ylim(c(0,1))+theme_bw(base_size = 18)+ggtitle(sample_group)
plots_list[[chrName]] <- p+guides(fill = "none" )
bins_pvals_list[[chrName]] <- unlist(bins_pvals)
}
mChrThresPlots <- marrangeGrob(plots_list, nrow=3, ncol=2)
notitle_p <- lapply(plots_list, function(x) x+ggtitle("")+ylab("")+
theme(plot.margin = unit(c(0.00,0.00,-0.02,0.00), "cm")))
mChrThresPlots <- marrangeGrob(notitle_p, ncol=7,nrow=3,
left = textGrob(paste0("Haplotype state ratio",sample_group),
rot = 90,gp = gpar(fontsize=22)),
layout_matrix = matrix(c(1:19,NA,NA), 3, 7, TRUE),
right= " ")
print(mChrThresPlots)
message(sample_group, " number of bins with FDR < 0.05: ",
sum(p.adjust(unlist(bins_pvals_list),"fdr")<0.05))
}Male_KO number of bins with FDR < 0.05: 0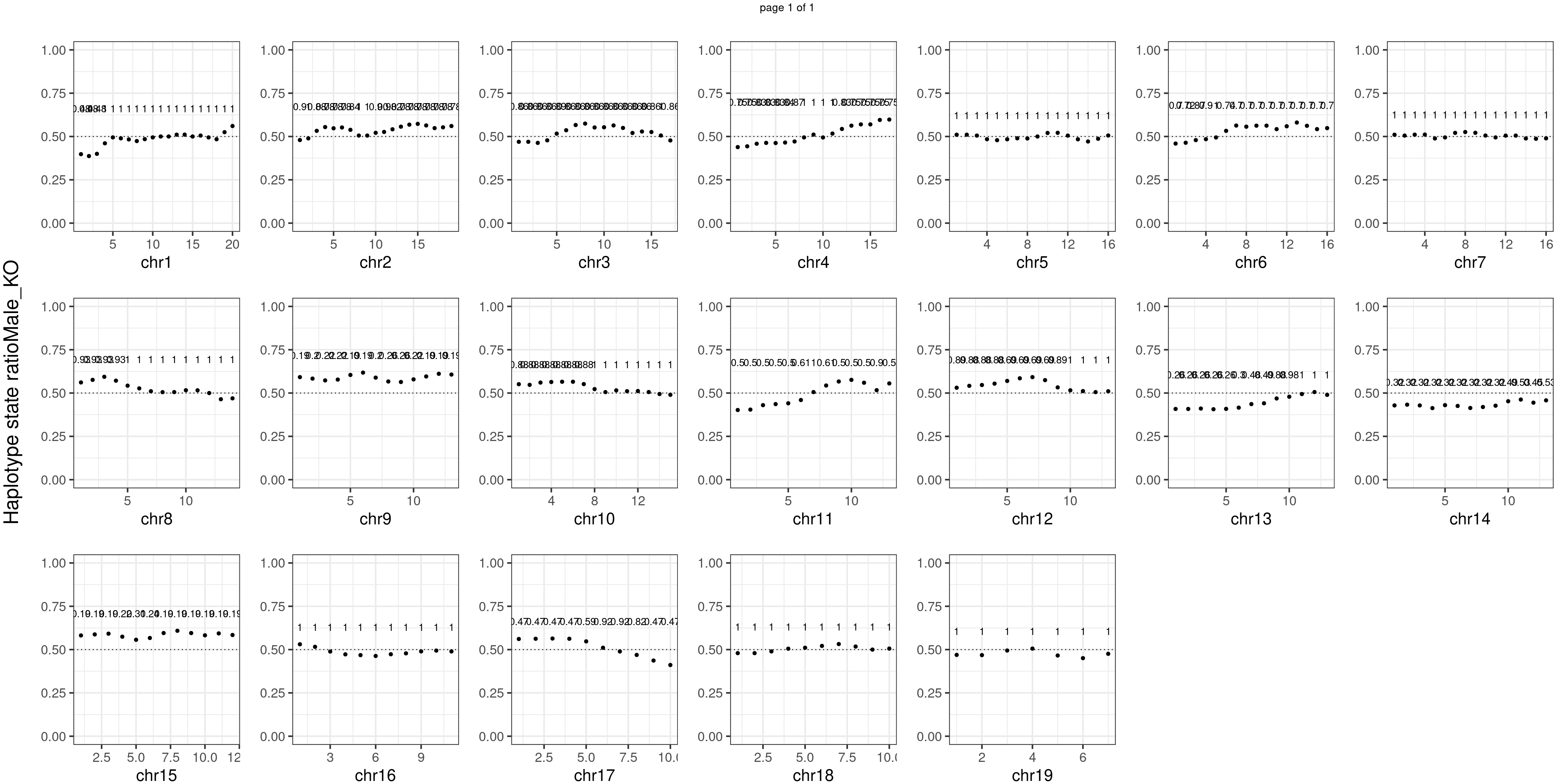
Female_KO number of bins with FDR < 0.05: 0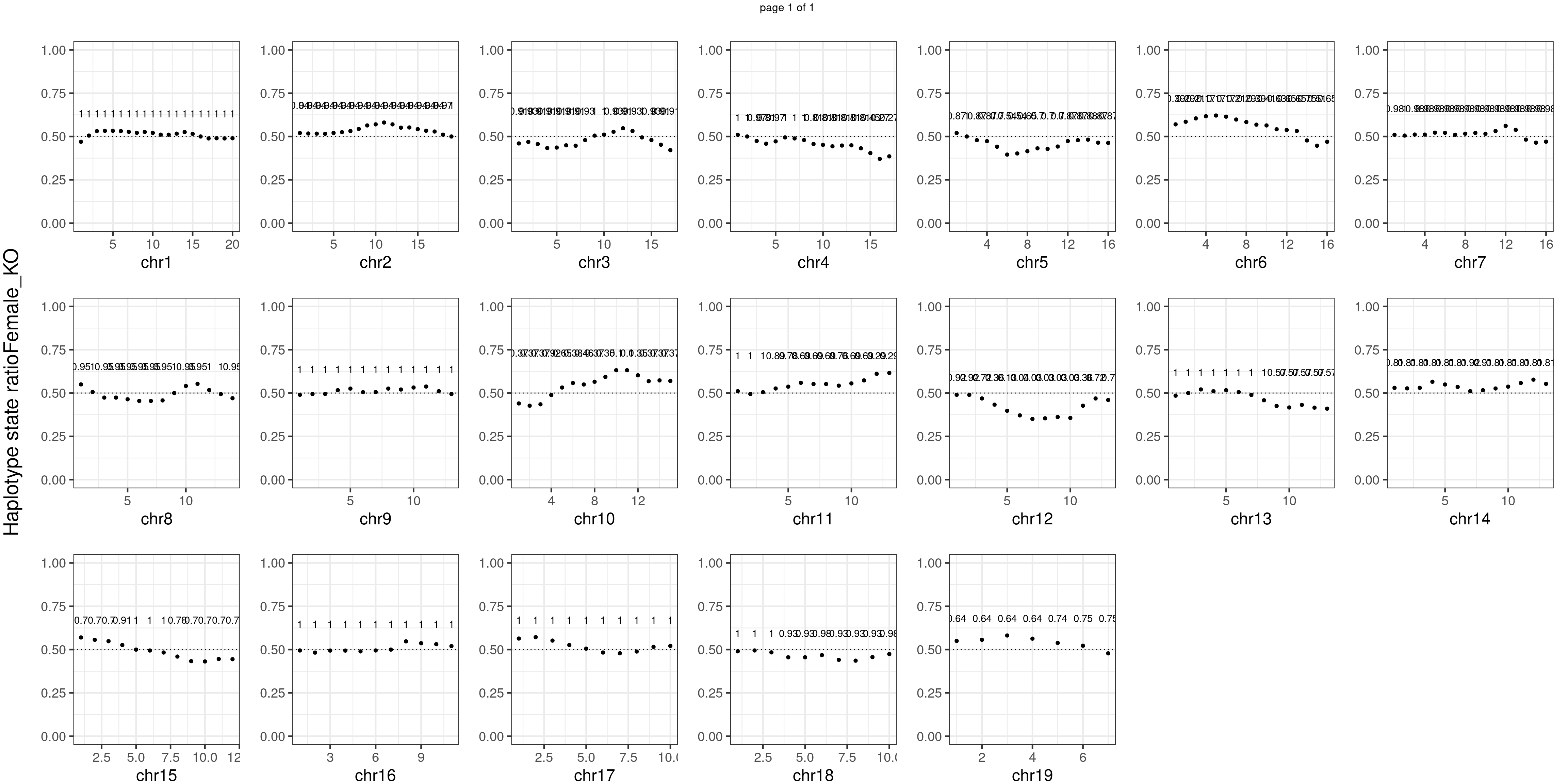
Female_WT number of bins with FDR < 0.05: 0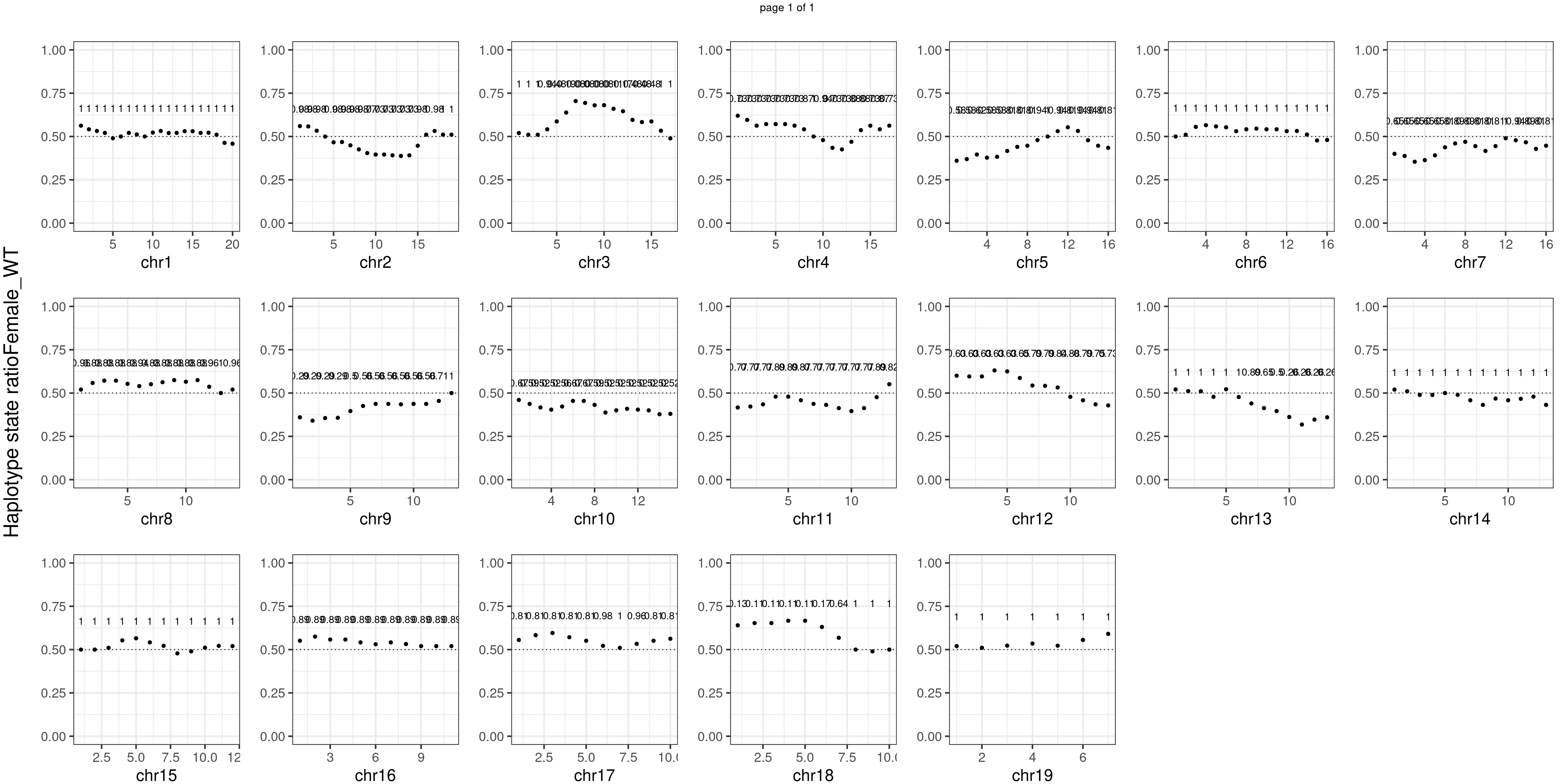
Female_HET number of bins with FDR < 0.05: 0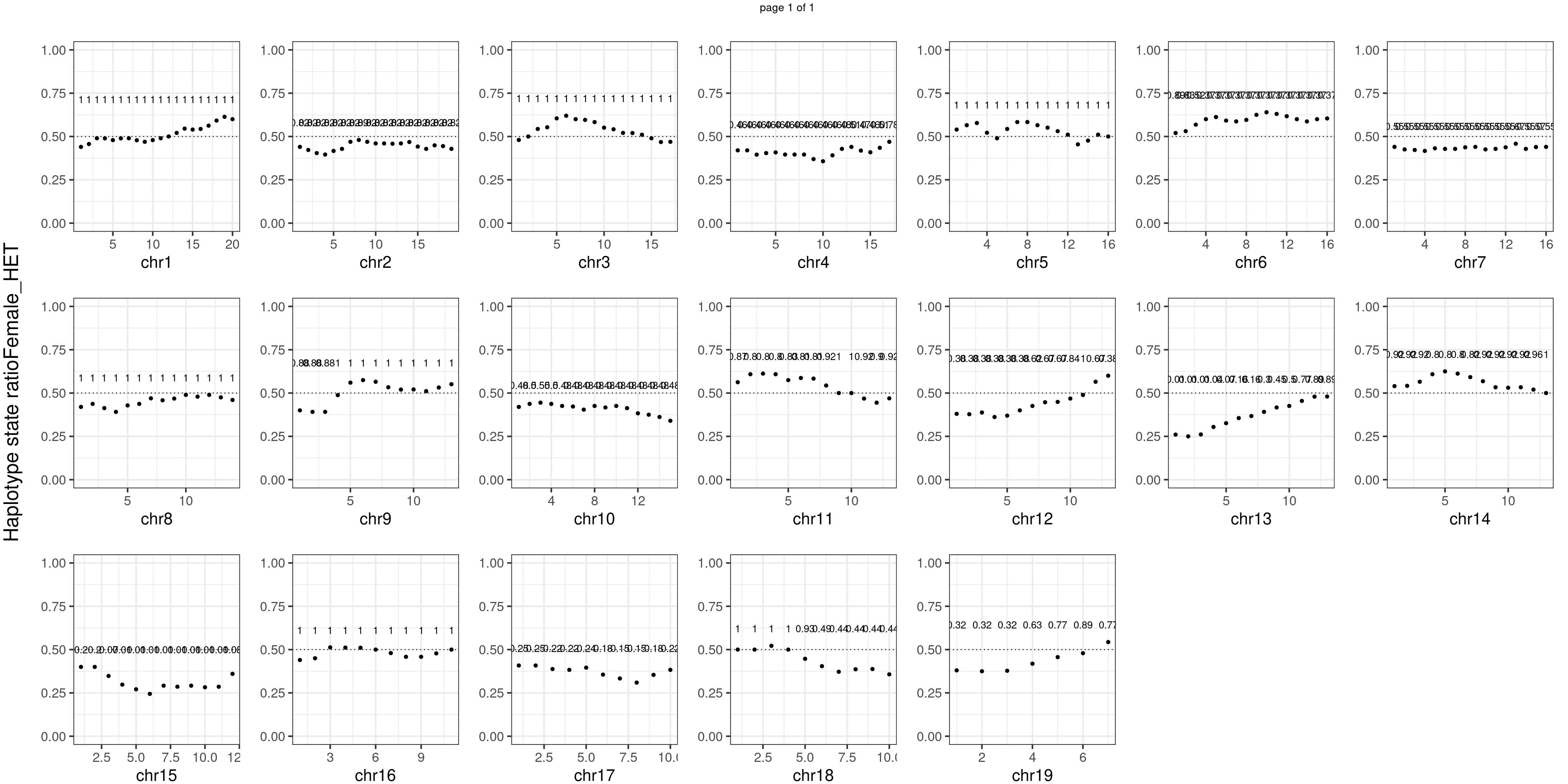
Male_WT number of bins with FDR < 0.05: 0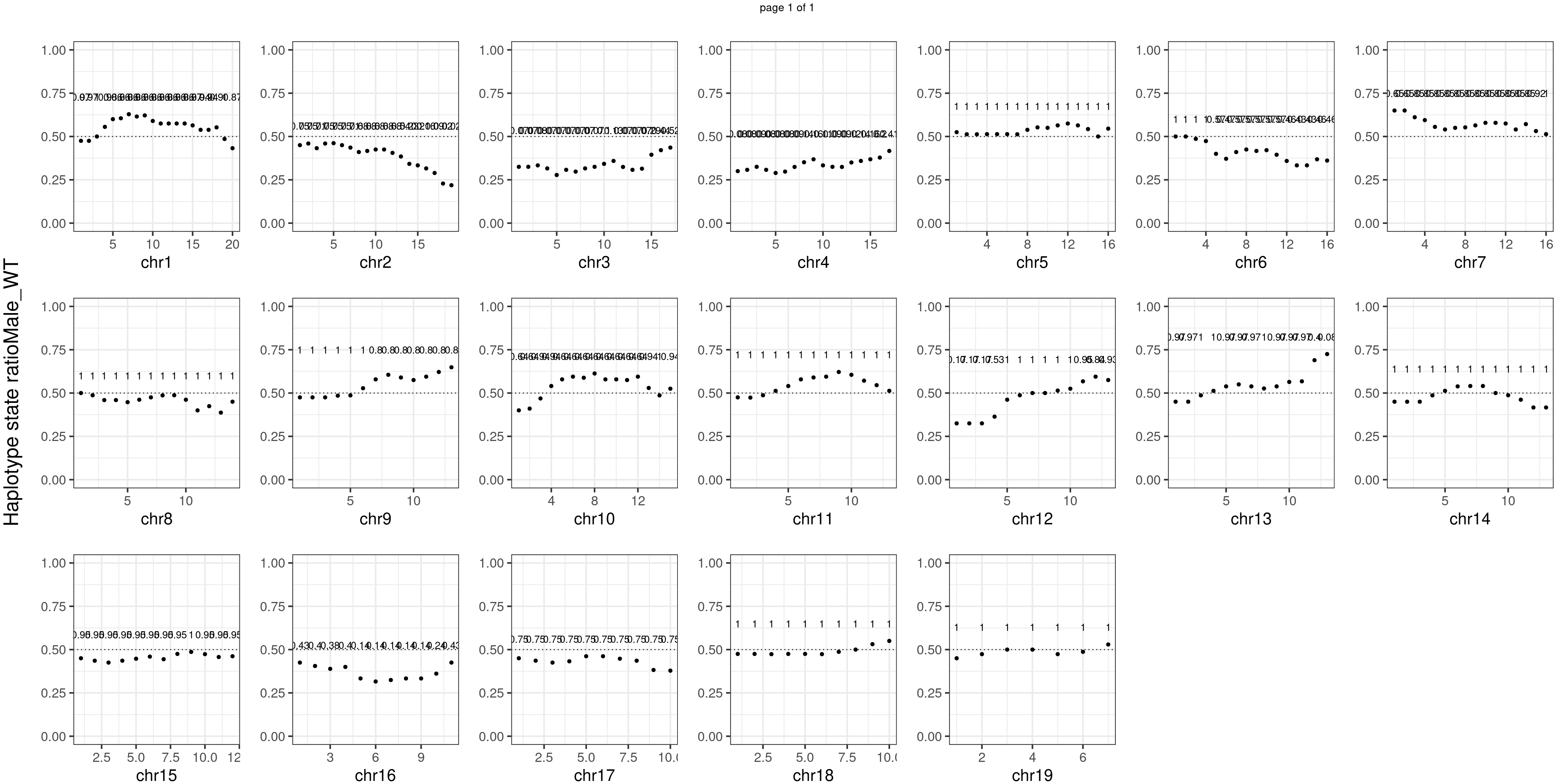
Male_HET number of bins with FDR < 0.05: 0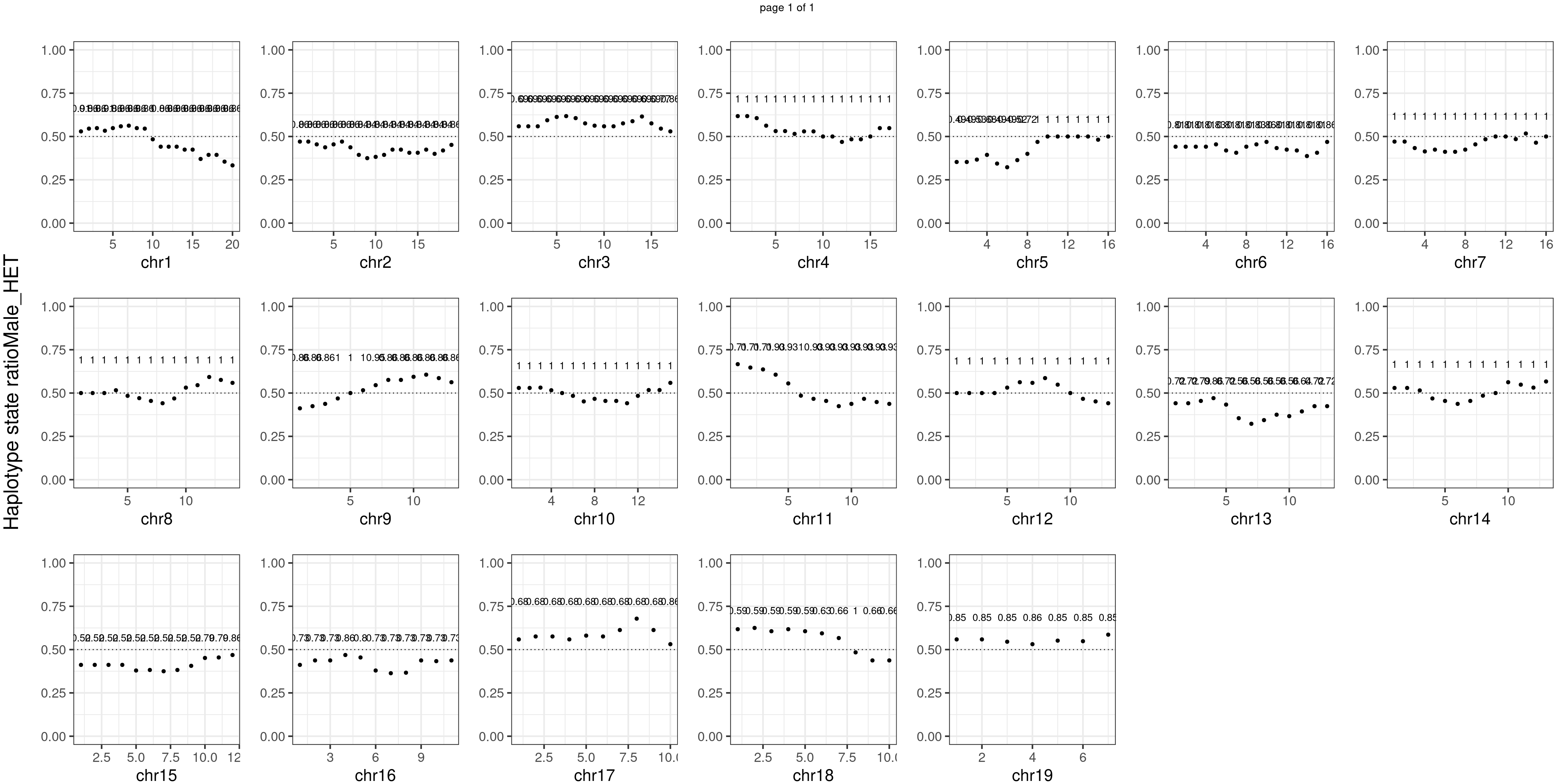
Conclusion
There is no apparent distorted segregation from the aggregated BC1F1 samples. Female_HET might worth having a closer look.
Mouse sex and genotype
The above grouping (Male_KO) was based on the genotype of BC1F1’s Fancm parent. Now we group by the mouse’s sex and genotype and check for female het specifically.
Find sex of the mice
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.1.0 (2021-05-18)
os Red Hat Enterprise Linux 8.4 (Ootpa)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-12-17
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
AnnotationDbi 1.54.1 2021-06-08 [1] Bioconductor
AnnotationFilter 1.16.0 2021-05-19 [1] Bioconductor
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.1.0)
backports 1.2.1 2020-12-09 [1] CRAN (R 4.1.0)
base64enc 0.1-3 2015-07-28 [1] CRAN (R 4.1.0)
Biobase 2.52.0 2021-05-19 [1] Bioconductor
BiocFileCache 2.0.0 2021-05-19 [1] Bioconductor
BiocGenerics * 0.38.0 2021-05-19 [1] Bioconductor
BiocIO 1.2.0 2021-05-19 [1] Bioconductor
BiocParallel 1.26.2 2021-08-22 [1] Bioconductor
biomaRt 2.48.3 2021-08-15 [1] Bioconductor
Biostrings 2.60.2 2021-08-05 [1] Bioconductor
biovizBase 1.40.0 2021-05-19 [1] Bioconductor
bit 4.0.4 2020-08-04 [1] CRAN (R 4.1.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.1.0)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.1.0)
blob 1.2.2 2021-07-23 [1] CRAN (R 4.1.0)
BSgenome 1.60.0 2021-05-19 [1] Bioconductor
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.1.0)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.0)
checkmate 2.0.0 2020-02-06 [1] CRAN (R 4.1.0)
circlize 0.4.13 2021-06-09 [1] CRAN (R 4.1.0)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.1.0)
cluster 2.1.2 2021-04-17 [2] CRAN (R 4.1.0)
codetools 0.2-18 2020-11-04 [2] CRAN (R 4.1.0)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.1.0)
comapr * 0.99.37 2021-11-28 [1] Github (ruqianl/comapr@aad1b6a)
crayon 1.4.1 2021-02-08 [1] CRAN (R 4.1.0)
curl 4.3.2 2021-06-23 [1] CRAN (R 4.1.0)
data.table 1.14.2 2021-09-27 [1] CRAN (R 4.1.0)
DBI 1.1.1 2021-01-15 [1] CRAN (R 4.1.0)
dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.1.0)
DelayedArray 0.18.0 2021-05-19 [1] Bioconductor
desc 1.4.0 2021-09-28 [1] CRAN (R 4.1.0)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.1.0)
dichromat 2.0-0 2013-01-24 [1] CRAN (R 4.1.0)
digest 0.6.28 2021-09-23 [1] CRAN (R 4.1.0)
dplyr * 1.0.7 2021-06-18 [1] CRAN (R 4.1.0)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
ensembldb 2.16.4 2021-08-05 [1] Bioconductor
evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.0)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.1.0)
farver 2.1.0 2021-02-28 [1] CRAN (R 4.1.0)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
filelock 1.0.2 2018-10-05 [1] CRAN (R 4.1.0)
foreach 1.5.1 2020-10-15 [1] CRAN (R 4.1.0)
foreign 0.8-81 2020-12-22 [2] CRAN (R 4.1.0)
Formula 1.2-4 2020-10-16 [1] CRAN (R 4.1.0)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.1.0)
generics 0.1.0 2020-10-31 [1] CRAN (R 4.1.0)
GenomeInfoDb * 1.28.4 2021-09-05 [1] Bioconductor
GenomeInfoDbData 1.2.6 2021-09-30 [1] Bioconductor
GenomicAlignments 1.28.0 2021-05-19 [1] Bioconductor
GenomicFeatures 1.44.2 2021-08-26 [1] Bioconductor
GenomicRanges * 1.44.0 2021-05-19 [1] Bioconductor
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.1.0)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.1.0)
GlobalOptions 0.1.2 2020-06-10 [1] CRAN (R 4.1.0)
glue 1.4.2 2020-08-27 [1] CRAN (R 4.1.0)
gridExtra * 2.3 2017-09-09 [1] CRAN (R 4.1.0)
gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.0)
Gviz 1.36.2 2021-07-04 [1] Bioconductor
highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
Hmisc 4.5-0 2021-02-28 [1] CRAN (R 4.1.0)
hms 1.1.1 2021-09-26 [1] CRAN (R 4.1.0)
htmlTable 2.2.1 2021-05-18 [1] CRAN (R 4.1.0)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
htmlwidgets 1.5.4 2021-09-08 [1] CRAN (R 4.1.0)
httpuv 1.6.3 2021-09-09 [1] CRAN (R 4.1.0)
httr 1.4.2 2020-07-20 [1] CRAN (R 4.1.0)
IRanges * 2.26.0 2021-05-19 [1] Bioconductor
iterators 1.0.13 2020-10-15 [1] CRAN (R 4.1.0)
jpeg 0.1-9 2021-07-24 [1] CRAN (R 4.1.0)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.1.0)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.0)
KEGGREST 1.32.0 2021-05-19 [1] Bioconductor
knitr 1.36 2021-09-29 [1] CRAN (R 4.1.0)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.1.0)
later 1.3.0 2021-08-18 [1] CRAN (R 4.1.0)
lattice 0.20-44 2021-05-02 [2] CRAN (R 4.1.0)
latticeExtra 0.6-29 2019-12-19 [1] CRAN (R 4.1.0)
lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.1.0)
lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.1.0)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.0)
Matrix 1.3-3 2021-05-04 [2] CRAN (R 4.1.0)
MatrixGenerics 1.4.3 2021-08-26 [1] Bioconductor
matrixStats 0.61.0 2021-09-17 [1] CRAN (R 4.1.0)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.1.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.0)
nnet 7.3-16 2021-05-03 [2] CRAN (R 4.1.0)
pillar 1.6.3 2021-09-26 [1] CRAN (R 4.1.0)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.1.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
pkgload 1.2.2 2021-09-11 [1] CRAN (R 4.1.0)
plotly 4.9.4.1 2021-06-18 [1] CRAN (R 4.1.0)
plyr 1.8.6 2020-03-03 [1] CRAN (R 4.1.0)
png 0.1-7 2013-12-03 [1] CRAN (R 4.1.0)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.0)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.0)
progress 1.2.2 2019-05-16 [1] CRAN (R 4.1.0)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.1.0)
ProtGenerics 1.24.0 2021-05-19 [1] Bioconductor
ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.0)
purrr 0.3.4 2020-04-17 [1] CRAN (R 4.1.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.1.0)
RColorBrewer 1.1-2 2014-12-07 [1] CRAN (R 4.1.0)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.1.0)
RCurl 1.98-1.5 2021-09-17 [1] CRAN (R 4.1.0)
remotes 2.4.1 2021-09-29 [1] CRAN (R 4.1.0)
reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.1.0)
restfulr 0.0.13 2017-08-06 [1] CRAN (R 4.1.0)
rjson 0.2.20 2018-06-08 [1] CRAN (R 4.1.0)
rlang 0.4.11 2021-04-30 [1] CRAN (R 4.1.0)
rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.1.0)
rpart 4.1-15 2019-04-12 [2] CRAN (R 4.1.0)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.0)
Rsamtools 2.8.0 2021-05-19 [1] Bioconductor
RSQLite 2.2.8 2021-08-21 [1] CRAN (R 4.1.0)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
rtracklayer 1.52.1 2021-08-15 [1] Bioconductor
S4Vectors * 0.30.1 2021-09-26 [1] Bioconductor
scales 1.1.1 2020-05-11 [1] CRAN (R 4.1.0)
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.1.0)
shape 1.4.6 2021-05-19 [1] CRAN (R 4.1.0)
stringi 1.7.4 2021-08-25 [1] CRAN (R 4.1.0)
stringr 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
SummarizedExperiment 1.22.0 2021-05-19 [1] Bioconductor
survival 3.2-11 2021-04-26 [2] CRAN (R 4.1.0)
testthat 3.0.4 2021-07-01 [1] CRAN (R 4.1.0)
tibble 3.1.4 2021-08-25 [1] CRAN (R 4.1.0)
tidyr 1.1.4 2021-09-27 [1] CRAN (R 4.1.0)
tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.1.0)
usethis 2.0.1 2021-02-10 [1] CRAN (R 4.1.0)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.1.0)
VariantAnnotation 1.38.0 2021-05-19 [1] Bioconductor
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.1.0)
viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.1.0)
whisker 0.4 2019-08-28 [1] CRAN (R 4.1.0)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.1.0)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.1.0)
xfun 0.26 2021-09-14 [1] CRAN (R 4.1.0)
XML 3.99-0.8 2021-09-17 [1] CRAN (R 4.1.0)
xml2 1.3.2 2020-04-23 [1] CRAN (R 4.1.0)
XVector 0.32.0 2021-05-19 [1] Bioconductor
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.0)
zlibbioc 1.38.0 2021-05-19 [1] Bioconductor
[1] /mnt/beegfs/mccarthy/scratch/general/rlyu/Software/R/Rlib/4.1.0/yeln
[2] /opt/R/4.1.0/lib/R/library