BAUH: MND scRNA-seq pilot 2: SoupX
C.B. Azodi
2021-09-29
Last updated: 2021-09-29
Checks: 6 1
Knit directory: BAUH_2020_MND-single-cell/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190102) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1728604. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/1-s2.0-S0002929720300781-main.pdf
Ignored: data/2103.11251.pdf
Ignored: data/3M-february-2018.txt
Ignored: data/737K-august-2016.txt
Ignored: data/STAR_index/
Ignored: data/STAR_output/
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.log
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.vcf.gz
Ignored: data/genome1k.chr22.log
Ignored: data/genome1k.chr22.recode.vcf
Ignored: data/s41588-018-0268-8.pdf
Ignored: data/tr2g_hs.tsv
Ignored: logs/
Ignored: output/CB-scRNAv31-GEX-lib01_QC_metadata.txt
Ignored: output/CB-scRNAv31-GEX-lib02_QC_metadata.txt
Ignored: output/pilot1_starsoloED/
Ignored: output/pilot2.1_gex/
Ignored: output/pilot2_HTO-2/
Ignored: output/pilot2_HTO/
Ignored: output/pilot2_gex_MAF01-152/
Ignored: output/pilot2_gex_MAF01/
Ignored: output/pilot2_gex_starsolo/
Ignored: output/pilot2_gex_starsoloED/
Ignored: output/pilot2_gex_starsoloED_GFP/
Ignored: references/SAindex/
Ignored: references/geno_test.vcf.gz
Untracked files:
Untracked: #14_venn_diagramm.png
Untracked: #14_venn_diagramm.png.2021-09-28_14-00-52.log
Untracked: #14_venn_diagramm.png.2021-09-28_14-01-05.log
Untracked: #14_venn_diagramm.png.2021-09-28_14-02-15.log
Untracked: #14_venn_diagramm.png.2021-09-28_14-02-22.log
Untracked: .snakemake/
Untracked: 2021-04-27_pilot2_nCells-per-donor.pdf
Untracked: 2021-08-03_pilot2_nCells-per-donor.pdf
Untracked: BAUH_2020_MND-single-cell.Rproj
Untracked: GRCh38_turboGFP-RFP_reference/
Untracked: Log.out
Untracked: Rplots.pdf
Untracked: code/run_soupX_2.R
Untracked: star-help.txt
Untracked: test/
Untracked: test_learn/
Untracked: test_maf01_notFiltered/
Untracked: test_maf05/
Untracked: test_noGeno/
Untracked: test_vireo/
Untracked: workflow/dropkick_get_ambient_and_hvgs.py
Unstaged changes:
Modified: analysis/2021-09-28_pilot2_SoupX.Rmd
Modified: analysis/2021-09-29_pilot2_VireoResults.Rmd
Modified: workflow/config_ambient_pilot2.1.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/2021-09-28_pilot2_SoupX.Rmd) and HTML (public/2021-09-28_pilot2_SoupX.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1728604 | cazodi | 2021-09-29 | add pilot1 analysis to workflowr |
#devtools::install_github("yanlinlin82/ggvenn")
suppressPackageStartupMessages({
library(ggplot2)
library(SoupX)
library(Seurat)
library(DropletUtils)
library(Matrix)
library(cowplot)
})
rerun <- FALSEFilter raw sce object
To include barcodes called as cells by either EmptyDroplet or dropkick
id <- "lib01"
out.raw <- paste0("output/pilot2.1_gex/02_EmptyDropDropkick/CB-scRNAv31-GEX-", id, "_outs/raw_feature_bc_matrix")
out.filt <- paste0("output/pilot2.1_gex/02_EmptyDropDropkick/CB-scRNAv31-GEX-", id, "_outs/filtered_feature_bc_matrix")
if (rerun) {
raw.dir <- paste0("output/pilot2.1_gex/01_cellranger/CB-scRNAv31-GEX-", id, "_S1/outs/raw_feature_bc_matrix/")
dk.file <- paste0("output/pilot2.1_gex/", id, "_EmptyDropDropkick-barcodes.tsv")
out <- paste0("output/pilot2.1_gex/02_EmptyDropDropkick/CB-scRNAv31-GEX-", id, "_outs/")
plot_out <- paste0("output/pilot2.1_gex/02_EmptyDropDropkick/CB-scRNAv31-GEX-", id, "_outs/plot_")
### Filter cellRanger results using dropkick cells ###
rawData <- Read10X(data.dir = raw.dir)
dkCells <- scan(dk.file, what = "character")
filtData <- rawData[, dkCells]
write10xCounts(out.filt, filtData, row.names(filtData),
gene.symbol=row.names(filtData), barcodes=colnames(filtData))
write10xCounts(out.raw, rawData, row.names(rawData),
gene.symbol=row.names(rawData), barcodes=colnames(rawData))
}Clustering
### Seurat: find HVGs
sc.filt <- Seurat::Read10X(data.dir = out.filt)
sc.filt <- CreateSeuratObject(counts = sc.filt)
sc.filt <- FindVariableFeatures(sc.filt, nfeatures = 2000)
varInfo <- HVFInfo(object = sc.filt)
hvgs5pThresh <- quantile(varInfo$variance.standardized, 0.95)
hvgs5p <- row.names(varInfo[varInfo$variance.standardized >= hvgs5pThresh, ])
### Seurat: Get UMAP embeddings
sc.filt <- RunUMAP(object = sc.filt, features=hvgs5p)16:27:05 UMAP embedding parameters a = 0.9922 b = 1.11216:27:06 Read 13127 rows and found 2920 numeric columns16:27:06 Using Annoy for neighbor search, n_neighbors = 3016:27:06 Building Annoy index with metric = cosine, n_trees = 500% 10 20 30 40 50 60 70 80 90 100%[----|----|----|----|----|----|----|----|----|----|**************************************************|
16:27:20 Writing NN index file to temp file /tmp/RtmpUa6CVY/file148a691e32ff
16:27:20 Searching Annoy index using 1 thread, search_k = 3000
16:29:30 Annoy recall = 100%
16:29:31 Commencing smooth kNN distance calibration using 1 thread
16:29:32 Initializing from normalized Laplacian + noise
Spectral initialization failed to converge, using random initialization instead
16:29:32 Commencing optimization for 200 epochs, with 733454 positive edges
16:29:39 Optimization finishedsc.umap <- Embeddings(object = sc.filt, reduction = "umap")
### Seurat: Generate clusters for cells
sc.filt <- ScaleData(sc.filt)Centering and scaling data matrixsc.filt <- RunPCA(sc.filt, features=hvgs5p)PC_ 1
Positive: FBP2, AC090819.1, RN7SL395P, MED15P8, AL645949.2, RNU5E-4P, DNAH17-AS1, LINC01425, SIGLEC1, RAB5CP1
GAPLINC, SPRR2A, AP001271.2, LUZP4P1, AMTN, AL117328.2, AL133343.2, ADH1B, RPL12P30, CASP12
RPL35P9, AC044802.2, FAM35CP, AP000851.2, AL645820.1, AC010969.1, PSPC1-AS2, LINC01291, AL137849.1, AC011131.1
Negative: HSP90AB1, PTMA, CALM2, TERF2IP, CFL1, STMN1, SRP14, DNAJA1, H3F3A, NCL
CD24, MORF4L1, CALM1, SET, MARCKSL1, H3F3B, MLLT11, ACTG1, SUMO2, RTN4
XRCC5, HSPA8, AASDHPPT, NUCKS1, EID1, HNRNPK, KHDRBS1, ATXN7L3B, YWHAB, CSDE1
PC_ 2
Positive: STMN2, GAP43, TUBA1B, NEFL, RCAN2, BASP1, TUBB2B, TUBA1A, CALM2, TUBB4A
TUBB, ACTB, DCX, PRKAR2B, ACTG1, FGF13, CALM1, CFL1, RAB3C, YWHAE
STMN1, NAP1L5, MAPT, DOK6, TSHZ1, MLLT11, ALDOC, CCNI, YWHAQ, SPINK6
Negative: AL590326.1, AC010331.1, AC099791.2, AC110285.6, AC092171.4, KMT2E-AS1, AC132192.2, AL139089.1, LINC01089, AC008403.3
AC124016.1, AC233280.1, AC087239.1, AC010997.5, FBXL8, AL359504.2, AC068205.2, HOXA-AS2, SNHG3, AC005837.3
MATN4, MPZ, AP001412.1, TOB1-AS1, RNF165, TCTE3, AC013731.1, AC023908.3, ZNF236-DT, CAPN10-DT
PC_ 3
Positive: ETV5, FZD2, GSN, COL18A1, GLIS3, IGFBP7, LAMC1, RBPMS, COL5A2, COL3A1
RRBP1, MYOF, CALD1, CFI, COL1A1, LRP10, PDGFRB, AHNAK, FBLN1, SLC12A4
COL16A1, FN1, TGFBI, FKBP10, HSPG2, COL4A1, WLS, LCAT, EPHB4, COL11A1
Negative: AC110285.6, RNF165, AC099791.2, AL590326.1, AC010331.1, AC092171.4, AC008403.3, AC068205.2, AL139089.1, LINC01089
AC010997.5, AC233280.1, AC132192.2, AC124016.1, HOXA-AS2, TOB1-AS1, KMT2E-AS1, AC087239.1, FBXL8, MPZ
MATN4, AP001412.1, AC013731.1, TCTE3, TMEM240, AC005837.3, AC023908.3, AL359504.2, SNHG3, ZNF236-DT
PC_ 4
Positive: HOXA5, CRABP1, HOTAIRM1, HOXB8, LHX1-DT, IRX3, HOXA1, ZNF703, ONECUT1, LMO4
HOXA7, LINC01116, ZNF22, CBX5, CHMP2B, DBI, IDI1, NAP1L1, SYT6, STXBP6
ACAT2, MARCKSL1, POU3F1, MARCKS, CCNI, DDIT3, TSPYL4, PEA15, HBD, LRRC61
Negative: ITGA2, LIFR, HLA-A, CRHBP, HLA-B, HOXC10, HLA-C, OPCML, TurboGFP, RSPO2
LHX9, CLMP, FOXP1, ZBTB7C, NRP2, EBF1, ETV1, B2M, PRPH, GABRG1
NTRK3, NFIA, NFIB, PITX2, HOXA10, ASAP1, SPINK6, MPPED2, A2M, SV2C
PC_ 5
Positive: MALAT1, MEIS2, ZFHX3, MEG3, BCL11A, CRABP1, GRIN2B, NCAM1, ONECUT1, SLC35F1
CLSTN2, EBF3, CRIM1, BNC2, SEMA5A, COL18A1, HIPK2, FLNA, XIST, MYCBP2
PBX1, IQGAP1, LHX1-DT, CMIP, KIDINS220, ESRRG, SYT1, BACH2, ENAH, DYNC1H1
Negative: HIST1H4H, TurboGFP, HIST1H1C, B2M, HIST1H2AC, PPP1R17, H1F0, NES, HIST2H2BE, ACTA1
HOXC10, RAB11FIP1, S100A6, S100A10, MAP1LC3B, H2AFJ, HOXA10, ISG15, HLA-B, GADD45A
GYPC, RPS27L, ATF3, RHOC, HIST1H2BG, ZC3HAV1, DDIT3, CKS2, CEBPB, HLA-C sc.filt <- FindNeighbors(sc.filt, reduction = "pca", dims = 1:30)Computing nearest neighbor graph
Computing SNNsc.filt <- FindClusters(object = sc.filt)Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 13127
Number of edges: 398813
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8083
Number of communities: 11
Elapsed time: 1 secondssc.clusters <- sc.filt@meta.data
head(sc.clusters) orig.ident nCount_RNA nFeature_RNA RNA_snn_res.0.8
AAACCCAAGAGCAAGA-1 SeuratProject 962 587 1
AAACCCAAGTTGCCTA-1 SeuratProject 1028 652 1
AAACCCACACACTGGC-1 SeuratProject 5210 2886 7
AAACCCACACATGAAA-1 SeuratProject 1168 725 1
AAACCCACAGATCCTA-1 SeuratProject 2286 1187 0
AAACCCAGTCGAGTTT-1 SeuratProject 1443 814 0
seurat_clusters
AAACCCAAGAGCAAGA-1 1
AAACCCAAGTTGCCTA-1 1
AAACCCACACACTGGC-1 7
AAACCCACACATGAAA-1 1
AAACCCACAGATCCTA-1 0
AAACCCAGTCGAGTTT-1 0Ambient read correction
Use SoupX to model ambient reads and correct counts.
### Load data into SoupX object and add Seurat info ###
toc = Seurat::Read10X(data.dir = out.filt)
tod = Seurat::Read10X(data.dir = out.raw)
sc = SoupChannel(tod, toc)
sc <- SoupX::setClusters(sc, sc.clusters[colnames(sc$toc), c("seurat_clusters")])
sc <- setDR(sc, sc.umap[colnames(sc$toc), c("UMAP_1", "UMAP_2")])
sc <- autoEstCont(sc, forceAccept = TRUE)9080 genes passed tf-idf cut-off and 5244 soup quantile filter. Taking the top 100.Using 488 independent estimates of rho.Estimated global rho of 0.76Extremely high contamination estimated (0.76). This likely represents a failure in estimating the contamination fraction. Set forceAccept=TRUE to proceed with this value.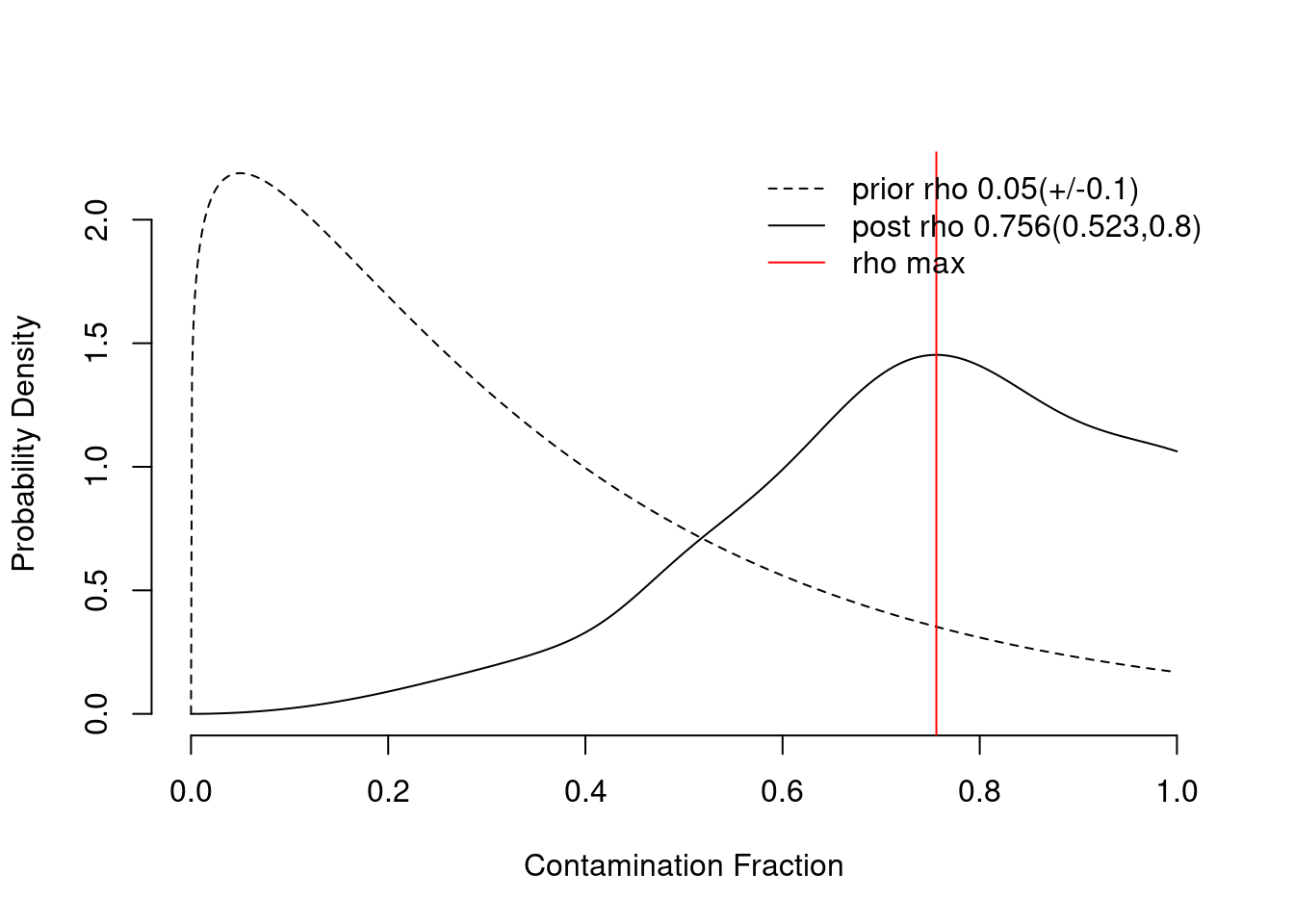
Sanity checks
For model estimated cluster marker genes:
plotMarkerDistribution(sc)No gene lists provided, attempting to find and plot cluster marker genes.Found 9080 marker genes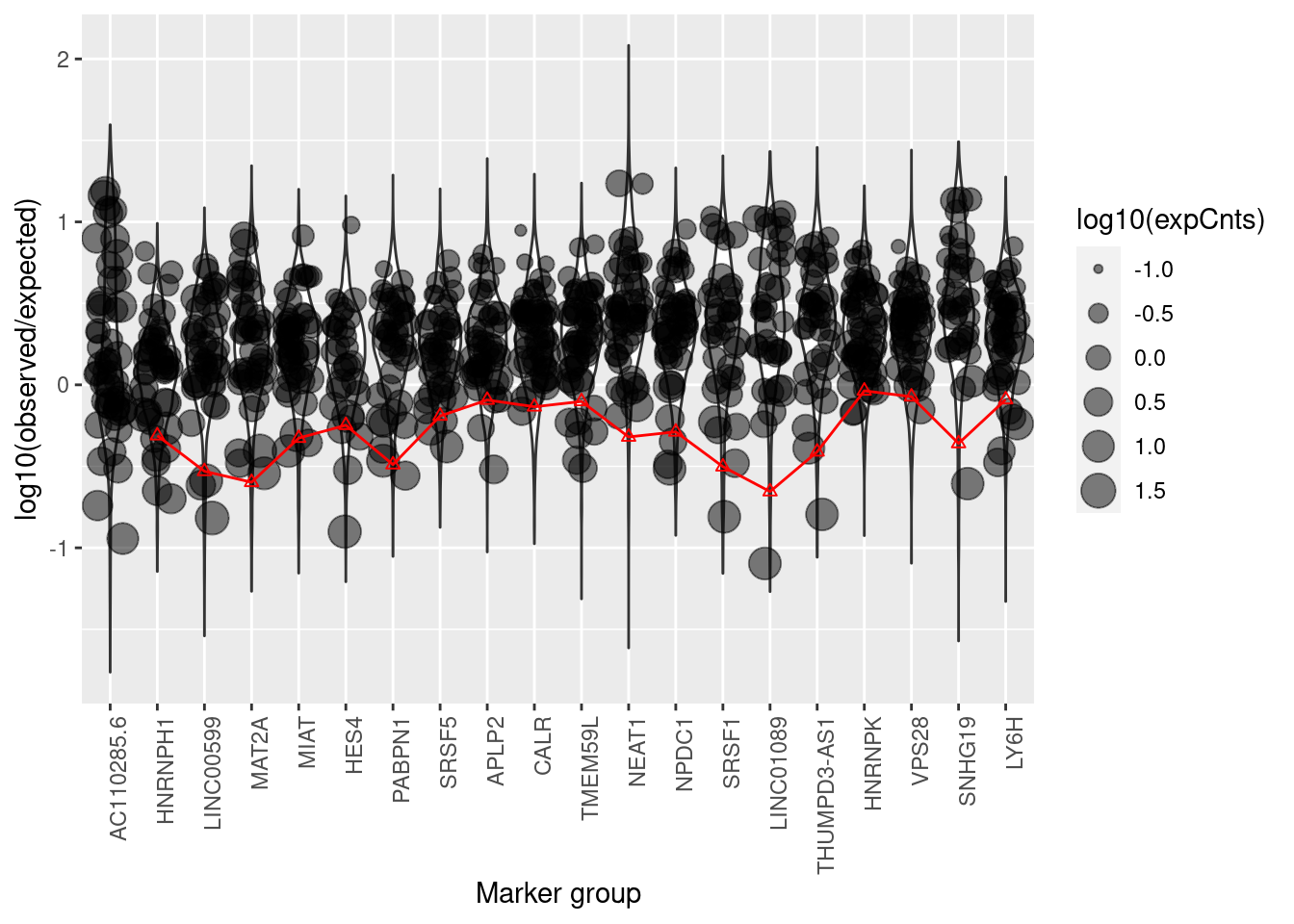
The distribution of log10 ratios of observed to expected counts if the cell contained nothing but soup. The red line shows the global estimate (i.e., assuming the same contamination fraction for all cells) of the contamination fraction using just that gene.
Marker gene sanity check
As another sanity check, we can look to see if motor neuron marker genes are present at greater than expected levels in cells. Marker genes suggested by Chris are: CHAT, IsL1, MNX1
chat <- plotMarkerMap(sc, "CHAT")
isl1 <- plotMarkerMap(sc, "ISL1")
mnx1 <- plotMarkerMap(sc, "MNX1")
all <- plotMarkerMap(sc, c("CHAT", "ISL1", "MNX1"))
plot_grid(chat, isl1, mnx1, all, labels="auto")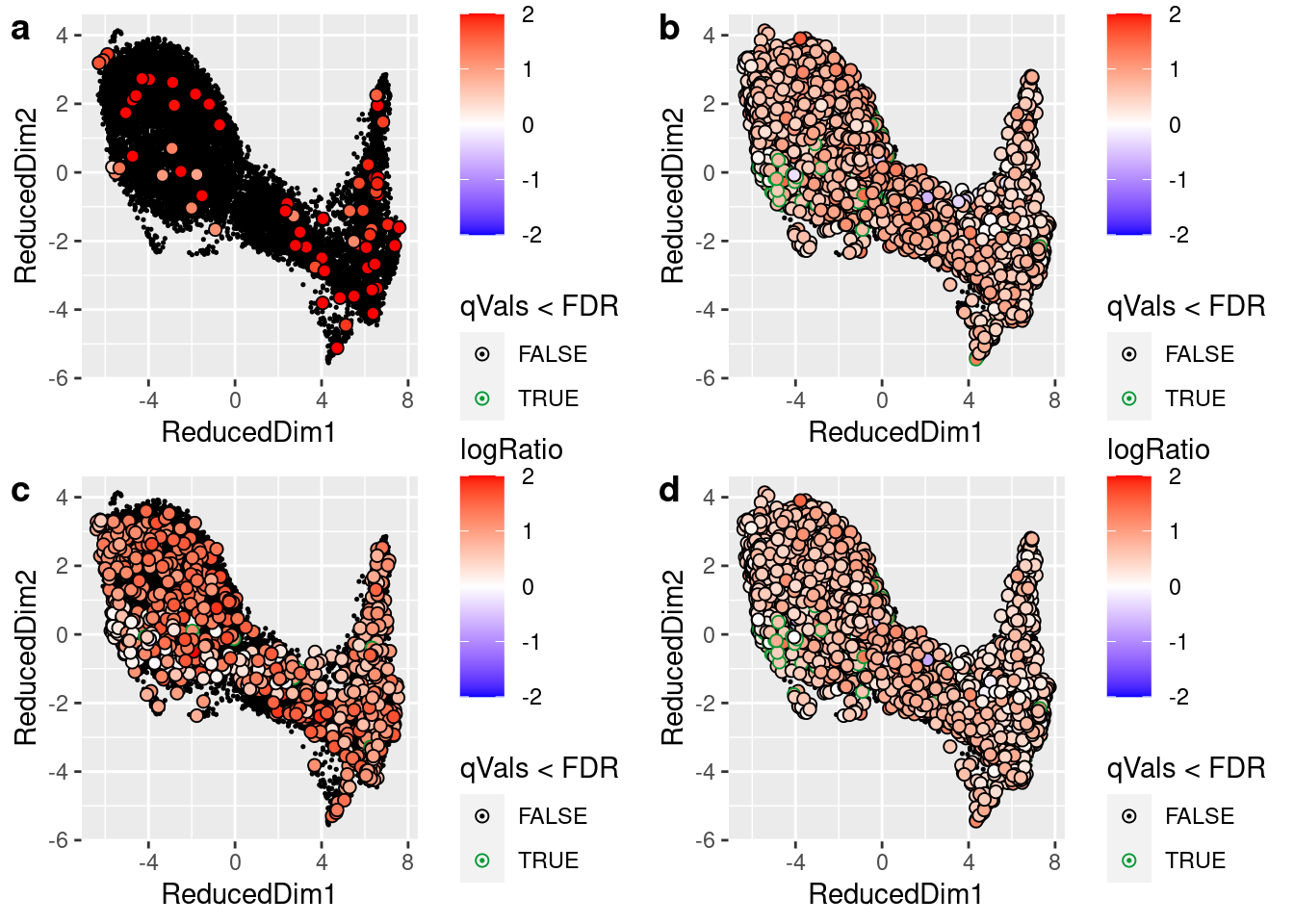
The ratio of observed counts to the expected counts if we assumed that cell contained nothing but soup for motor neuron marker genes (a) CHAT, (b) ISL1, (c) MNX1, and (d) all three together.
De-novo count correction
out <- adjustCounts(sc)Expanding counts from 11 clusters to 13127 cells.plotChangeMap(sc, out, "MNX1")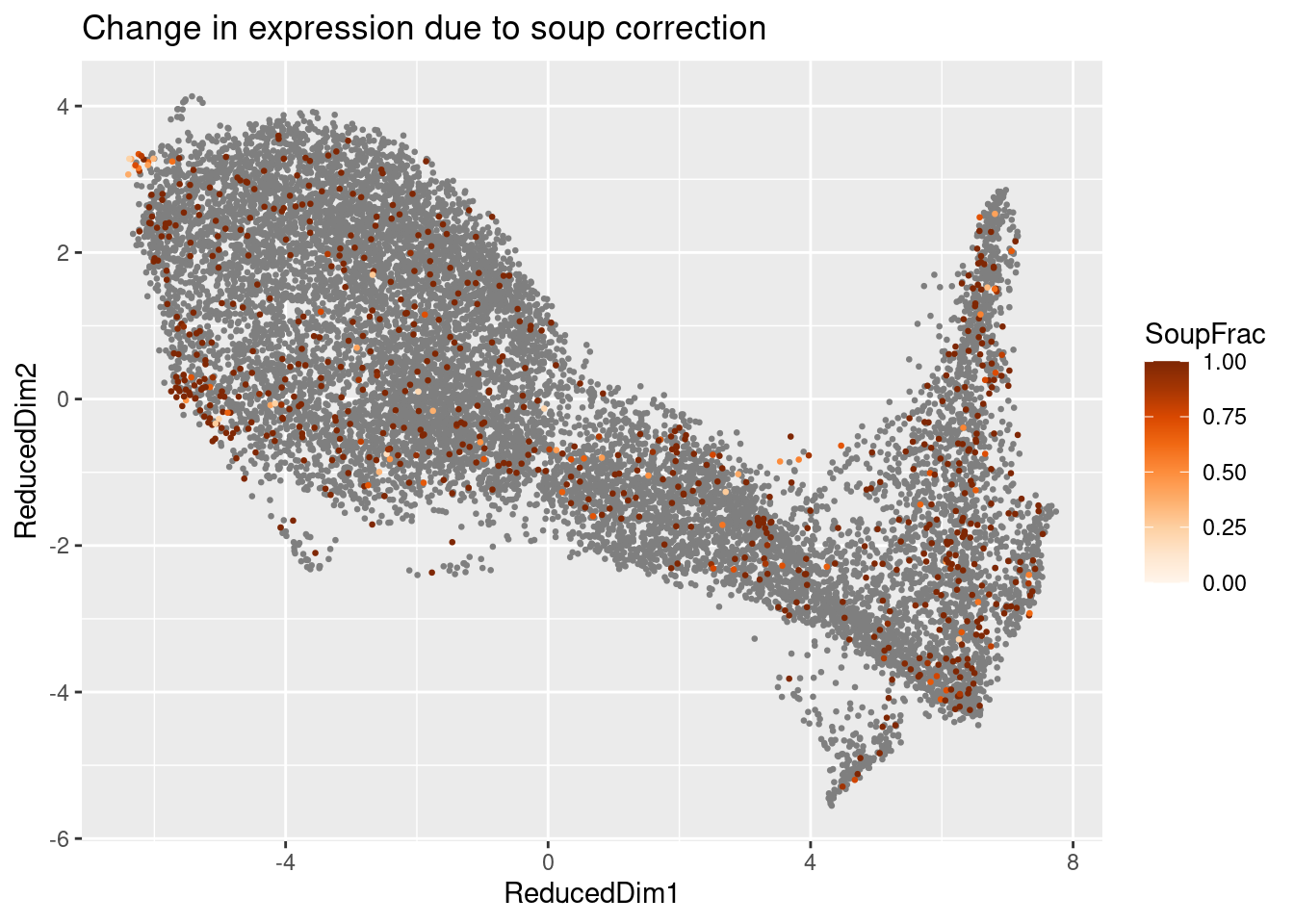
dk.dir <- "output/pilot2.1_gex/02_dropkick/CB-scRNAv31-GEX-lib01_S1/"
if(rerun) {
### SoupX corrected counts for dropkick called cells
sce <- read10xCounts("output/pilot2.1_gex/03_soupX/CB-scRNAv31-GEX-lib01/")
colnames(sce) <- sce$Barcode
### Raw cellranger counts for dropkick called cells
raw.dir <- "output/pilot2.1_gex/01_cellranger/CB-scRNAv31-GEX-lib01_S1/outs/raw_feature_bc_matrix/"
dkCells <- scan(paste0(dk.dir, "raw_feature_bc_matrix_dropkick_barcodes.txt"), what = "character")
rawData <- Seurat::Read10X(data.dir = raw.dir)
rawData <- Seurat::CreateSeuratObject(counts = rawData)
filtData <- rawData[, dkCells]
sce.dk <- Seurat::as.SingleCellExperiment(filtData)
assays(sce)$raw <- counts(sce.dk)
rm(sce.dk)
sce$raw_count <- colSums(as.matrix(assays(sce)$raw))
sce$SoupX_count <- colSums(as.matrix(assays(sce)$counts))
sce$ambient_drop_percent <- (sce$raw_count - sce$SoupX_count) / sce$raw_count
rm(rawData, filtData)
saveRDS(sce, paste0(dk.dir, "dk-soupX_filtered_sce.rds"))
} else{
sce <- readRDS(paste0(dk.dir, "dk-soupX_filtered_sce.rds"))
}
message(paste("SCE object contains", nrow(counts(sce)), "genes, and",
ncol(counts(sce)), "cells", sep=" "))SCE object contains 58397 genes, and 11019 cellsmessage("Summary of the percent of ambient reads dropped per cell:")Summary of the percent of ambient reads dropped per cell:summary(sce$ambient_drop_percent) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0439 0.1641 0.1792 0.1800 0.1951 0.3472
devtools::session_info()Registered S3 method overwritten by 'cli':
method from
print.boxx spatstat.geom─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.0.4 (2021-02-15)
os Rocky Linux 8.4 (Green Obsidian)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-09-29
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.0.2)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.2)
beachmat 2.6.4 2020-12-20 [1] Bioconductor
Biobase * 2.50.0 2020-10-27 [1] Bioconductor
BiocGenerics * 0.36.1 2021-04-16 [1] Bioconductor
BiocParallel 1.24.1 2020-11-06 [1] Bioconductor
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.0.4)
bslib 0.2.5.1 2021-05-18 [1] CRAN (R 4.0.4)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.4)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.0.4)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.0.4)
cluster 2.1.0 2019-06-19 [2] CRAN (R 4.0.4)
codetools 0.2-18 2020-11-04 [1] CRAN (R 4.0.2)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.0.4)
cowplot * 1.1.1 2020-12-30 [1] CRAN (R 4.0.4)
crayon 1.4.1 2021-02-08 [1] CRAN (R 4.0.4)
data.table 1.14.2 2021-09-27 [1] CRAN (R 4.0.4)
DBI 1.1.1 2021-01-15 [1] CRAN (R 4.0.4)
DelayedArray 0.16.3 2021-03-24 [1] Bioconductor
DelayedMatrixStats 1.12.3 2021-02-03 [1] Bioconductor
deldir 0.2-10 2021-02-16 [1] CRAN (R 4.0.4)
desc 1.3.0 2021-03-05 [1] CRAN (R 4.0.4)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.0.4)
digest 0.6.28 2021-09-23 [1] CRAN (R 4.0.4)
dplyr 1.0.7 2021-06-18 [1] CRAN (R 4.0.4)
dqrng 0.3.0 2021-05-01 [1] CRAN (R 4.0.4)
DropletUtils * 1.10.3 2021-02-02 [1] Bioconductor
edgeR 3.32.1 2021-01-14 [1] Bioconductor
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.0.4)
evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.2)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.0.4)
farver 2.1.0 2021-02-28 [1] CRAN (R 4.0.4)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.0.3)
fitdistrplus 1.1-5 2021-05-28 [1] CRAN (R 4.0.4)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
future 1.22.1 2021-08-25 [1] CRAN (R 4.0.4)
future.apply 1.8.1 2021-08-10 [1] CRAN (R 4.0.4)
generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
GenomeInfoDb * 1.26.7 2021-04-08 [1] Bioconductor
GenomeInfoDbData 1.2.4 2020-11-10 [1] Bioconductor
GenomicRanges * 1.42.0 2020-10-27 [1] Bioconductor
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.0.4)
ggrepel 0.9.1 2021-01-15 [1] CRAN (R 4.0.4)
ggridges 0.5.3 2021-01-08 [1] CRAN (R 4.0.4)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.0.4)
globals 0.14.0 2020-11-22 [1] CRAN (R 4.0.3)
glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
goftest 1.2-2 2019-12-02 [1] CRAN (R 4.0.3)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.0.2)
gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.2)
HDF5Array 1.18.1 2021-02-04 [1] Bioconductor
highr 0.9 2021-04-16 [1] CRAN (R 4.0.4)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.0.4)
htmlwidgets 1.5.4 2021-09-08 [1] CRAN (R 4.0.4)
httpuv 1.6.2 2021-08-18 [1] CRAN (R 4.0.4)
httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
ica 1.0-2 2018-05-24 [1] CRAN (R 4.0.3)
igraph 1.2.6 2020-10-06 [1] CRAN (R 4.0.4)
IRanges * 2.24.1 2020-12-12 [1] Bioconductor
irlba 2.3.3 2019-02-05 [1] CRAN (R 4.0.2)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.4)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.4)
KernSmooth 2.23-20 2021-05-03 [1] CRAN (R 4.0.4)
knitr 1.34 2021-09-09 [1] CRAN (R 4.0.4)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.0.2)
later 1.3.0 2021-08-18 [1] CRAN (R 4.0.4)
lattice 0.20-41 2020-04-02 [2] CRAN (R 4.0.4)
lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.0.2)
leiden 0.3.9 2021-07-27 [1] CRAN (R 4.0.4)
lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.0.4)
limma 3.46.0 2020-10-27 [1] Bioconductor
listenv 0.8.0 2019-12-05 [1] CRAN (R 4.0.2)
lmtest 0.9-38 2020-09-09 [1] CRAN (R 4.0.4)
locfit 1.5-9.4 2020-03-25 [1] CRAN (R 4.0.2)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.3)
MASS 7.3-54 2021-05-03 [1] CRAN (R 4.0.4)
Matrix * 1.3-4 2021-06-01 [1] CRAN (R 4.0.4)
MatrixGenerics * 1.2.1 2021-01-30 [1] Bioconductor
matrixStats * 0.60.0 2021-07-26 [1] CRAN (R 4.0.4)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.4)
mgcv 1.8-36 2021-06-01 [1] CRAN (R 4.0.4)
mime 0.11 2021-06-23 [1] CRAN (R 4.0.4)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.0.3)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.2)
nlme 3.1-152 2021-02-04 [1] CRAN (R 4.0.4)
parallelly 1.27.0 2021-07-19 [1] CRAN (R 4.0.4)
patchwork 1.1.1 2020-12-17 [1] CRAN (R 4.0.4)
pbapply 1.4-3 2020-08-18 [1] CRAN (R 4.0.3)
pillar 1.6.3 2021-09-26 [1] CRAN (R 4.0.4)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.4)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
pkgload 1.2.2 2021-09-11 [1] CRAN (R 4.0.4)
plotly 4.9.4.1 2021-06-18 [1] CRAN (R 4.0.4)
plyr 1.8.6 2020-03-03 [1] CRAN (R 4.0.2)
png 0.1-7 2013-12-03 [1] CRAN (R 4.0.4)
polyclip 1.10-0 2019-03-14 [1] CRAN (R 4.0.2)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.2)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.0.4)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.0.4)
ps 1.6.0 2021-02-28 [1] CRAN (R 4.0.4)
purrr 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
R.methodsS3 1.8.1 2020-08-26 [1] CRAN (R 4.0.2)
R.oo 1.24.0 2020-08-26 [1] CRAN (R 4.0.2)
R.utils 2.10.1 2020-08-26 [1] CRAN (R 4.0.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.0.4)
RANN 2.6.1 2019-01-08 [1] CRAN (R 4.0.3)
RColorBrewer 1.1-2 2014-12-07 [1] CRAN (R 4.0.2)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.0.4)
RcppAnnoy 0.0.19 2021-07-30 [1] CRAN (R 4.0.4)
RCurl 1.98-1.4 2021-08-17 [1] CRAN (R 4.0.4)
remotes 2.4.0 2021-06-02 [1] CRAN (R 4.0.4)
reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.0.2)
reticulate 1.20 2021-05-03 [1] CRAN (R 4.0.4)
rhdf5 2.34.0 2020-10-27 [1] Bioconductor
rhdf5filters 1.2.1 2021-05-03 [1] Bioconductor
Rhdf5lib 1.12.1 2021-01-26 [1] Bioconductor
rlang 0.4.11 2021-04-30 [1] CRAN (R 4.0.4)
rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.0.4)
ROCR 1.0-11 2020-05-02 [1] CRAN (R 4.0.2)
rpart 4.1-15 2019-04-12 [2] CRAN (R 4.0.4)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.3)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.3)
Rtsne 0.15 2018-11-10 [1] CRAN (R 4.0.4)
S4Vectors * 0.28.1 2020-12-09 [1] Bioconductor
sass 0.4.0 2021-05-12 [1] CRAN (R 4.0.4)
scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.2)
scattermore 0.7 2020-11-24 [1] CRAN (R 4.0.3)
sctransform 0.3.2 2020-12-16 [1] CRAN (R 4.0.4)
scuttle 1.0.4 2020-12-17 [1] Bioconductor
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
Seurat * 4.0.4 2021-08-20 [1] CRAN (R 4.0.4)
SeuratObject * 4.0.2 2021-06-09 [1] CRAN (R 4.0.4)
shiny 1.6.0 2021-01-25 [1] CRAN (R 4.0.4)
SingleCellExperiment * 1.12.0 2020-10-27 [1] Bioconductor
SoupX * 1.5.2 2021-05-17 [1] CRAN (R 4.0.4)
sparseMatrixStats 1.2.1 2021-02-02 [1] Bioconductor
spatstat.core 2.3-0 2021-07-16 [1] CRAN (R 4.0.4)
spatstat.data 2.1-0 2021-03-21 [1] CRAN (R 4.0.4)
spatstat.geom 2.2-2 2021-07-12 [1] CRAN (R 4.0.4)
spatstat.sparse 2.0-0 2021-03-16 [1] CRAN (R 4.0.4)
spatstat.utils 2.2-0 2021-06-14 [1] CRAN (R 4.0.4)
stringi 1.7.4 2021-08-25 [1] CRAN (R 4.0.4)
stringr 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
SummarizedExperiment * 1.20.0 2020-10-27 [1] Bioconductor
survival 3.2-13 2021-08-24 [1] CRAN (R 4.0.4)
tensor 1.5 2012-05-05 [1] CRAN (R 4.0.3)
testthat 3.0.4 2021-07-01 [1] CRAN (R 4.0.4)
tibble 3.1.4 2021-08-25 [1] CRAN (R 4.0.4)
tidyr 1.1.4 2021-09-27 [1] CRAN (R 4.0.4)
tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.0.4)
usethis 2.0.1 2021-02-10 [1] CRAN (R 4.0.4)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.4)
uwot 0.1.10 2020-12-15 [1] CRAN (R 4.0.4)
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.0.4)
viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.0.4)
whisker 0.4 2019-08-28 [1] CRAN (R 4.0.2)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.0.4)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.0.2)
xfun 0.26 2021-09-14 [1] CRAN (R 4.0.4)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.0.3)
XVector 0.30.0 2020-10-27 [1] Bioconductor
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
zlibbioc 1.36.0 2020-10-27 [1] Bioconductor
zoo 1.8-9 2021-03-09 [1] CRAN (R 4.0.4)
[1] /mnt/mcfiles/cazodi/R/x86_64-pc-linux-gnu-library/4.0
[2] /opt/R/4.0.4/lib/R/library