Pilot3: Cell calling with lenti barcodes
Christina Azodi
2022-03-02
Last updated: 2022-03-02
Checks: 5 2
Knit directory: BAUH_2020_MND-single-cell/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/03_keys/ | ../output/pilot3_Lenti/03_keys |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/ | ../output/pilot3.0_iPSC |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_MN/ | ../output/pilot3.0_MN |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3f19b81. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .cache/
Ignored: .config/
Ignored: .nv/
Ignored: .snakemake/
Ignored: BAUH_2020_MND-single-cell.Rproj
Ignored: GRCh38_turboGFP-RFP_reference/
Ignored: Homo_sapiens.GRCh38.turboGFP/
Ignored: Rplots.pdf
Ignored: data/1-s2.0-S0002929720300781-main.pdf
Ignored: data/2103.11251.pdf
Ignored: data/3M-february-2018.txt
Ignored: data/737K-august-2016.txt
Ignored: data/STAR_index/
Ignored: data/STAR_output/
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.log
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.vcf.gz
Ignored: data/genome1k.chr22.log
Ignored: data/genome1k.chr22.recode.vcf
Ignored: data/pilot3_aggr-experiments.csv
Ignored: data/pilot3_donors.txt
Ignored: data/s41588-018-0268-8.pdf
Ignored: data/tr2g_hs.tsv
Ignored: logs/
Ignored: output/2021-04-27_pilot2_nCells-per-donor.pdf
Ignored: output/2021-08-03_pilot2_nCells-per-donor.pdf
Ignored: output/CB-scRNAv31-GEX-lib01_QC_metadata.txt
Ignored: output/CB-scRNAv31-GEX-lib02_QC_metadata.txt
Ignored: output/pilot1_starsoloED/
Ignored: output/pilot2.1_gex/
Ignored: output/pilot2_HTO-2/
Ignored: output/pilot2_HTO/
Ignored: output/pilot2_gex_MAF01-152/
Ignored: output/pilot2_gex_MAF01/
Ignored: output/pilot2_gex_starsolo/
Ignored: output/pilot2_gex_starsoloED/
Ignored: output/pilot2_gex_starsoloED_GFP/
Ignored: output/pilot2_testing/
Ignored: output/pilot3.0/
Ignored: output/pilot3.0_MN/
Ignored: output/pilot3.0_captures-separate/
Ignored: output/pilot3.0_iPSC/
Ignored: output/pilot3_Lenti/
Ignored: references/Homo_sapiens.GRCh38.turboGFP.bed
Ignored: references/Homo_sapiens.GRCh38.turboGFP.fa
Ignored: references/Homo_sapiens.GRCh38.turboGFP.fa.fai
Ignored: references/Homo_sapiens.GRCh38.turboGFP.filtered.gtf
Ignored: references/Homo_sapiens.GRCh38.turboGFP.gtf
Ignored: references/SAindex/
Ignored: references/geno_test.vcf.gz
Ignored: references/pilot3/
Ignored: references/test
Ignored: references/turboGFP.fa
Ignored: references/turboGFP.gtf
Ignored: references/turboRFP.fa
Ignored: testupset_plot.pdf
Ignored: testvenn_diagram.pdf
Ignored: workflow/rules/
Untracked files:
Untracked: Capture5-GEX/
Untracked: __Capture5-GEX.mro
Untracked: analysis/2022-03-02_pilot3_ensemble-cellcalling.Rmd
Untracked: cellbender.dockerfile
Untracked: hwe1e-05_maf05_vcf_stats.txt
Untracked: hwe1e-05_vcf_stats.txt
Untracked: testbarcodes.txt
Untracked: testcombination_matrix.rds
Unstaged changes:
Modified: analysis/2022-01-07_pilot3_Cell-Calling-Comparison.Rmd
Modified: analysis/2022-02-28_pilot3_Cell-demultiplexing-Lenti.Rmd
Modified: analysis/2022-03-01_pilot3_cellbender.Rmd
Modified: analysis/2022-03-01_pilot3_dropkick.Rmd
Modified: code/get_barcodes_to_use.R
Modified: config/config_pilot3.0_MN.yml
Modified: config/config_pilot3.0_iPSC.yml
Modified: workflow/Snakefile
Modified: workflow/rules_cellcalling.smk
Modified: workflow/rules_demultiplexing.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/2022-02-28_pilot3_Cell-demultiplexing-Lenti.Rmd) and HTML (public/2022-02-28_pilot3_Cell-demultiplexing-Lenti.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a1fa048 | cazodi | 2022-03-01 | fix cellbenderGPU and update lenti downstream analysis |
| html | a1fa048 | cazodi | 2022-03-01 | fix cellbenderGPU and update lenti downstream analysis |
| Rmd | 409be72 | cazodi | 2022-02-28 | add strategy photos to workflowr |
| html | 409be72 | cazodi | 2022-02-28 | add strategy photos to workflowr |
| Rmd | 0f7f43b | cazodi | 2022-02-28 | lenti barcodes vs vireo results for ipscs |
| html | 0f7f43b | cazodi | 2022-02-28 | lenti barcodes vs vireo results for ipscs |
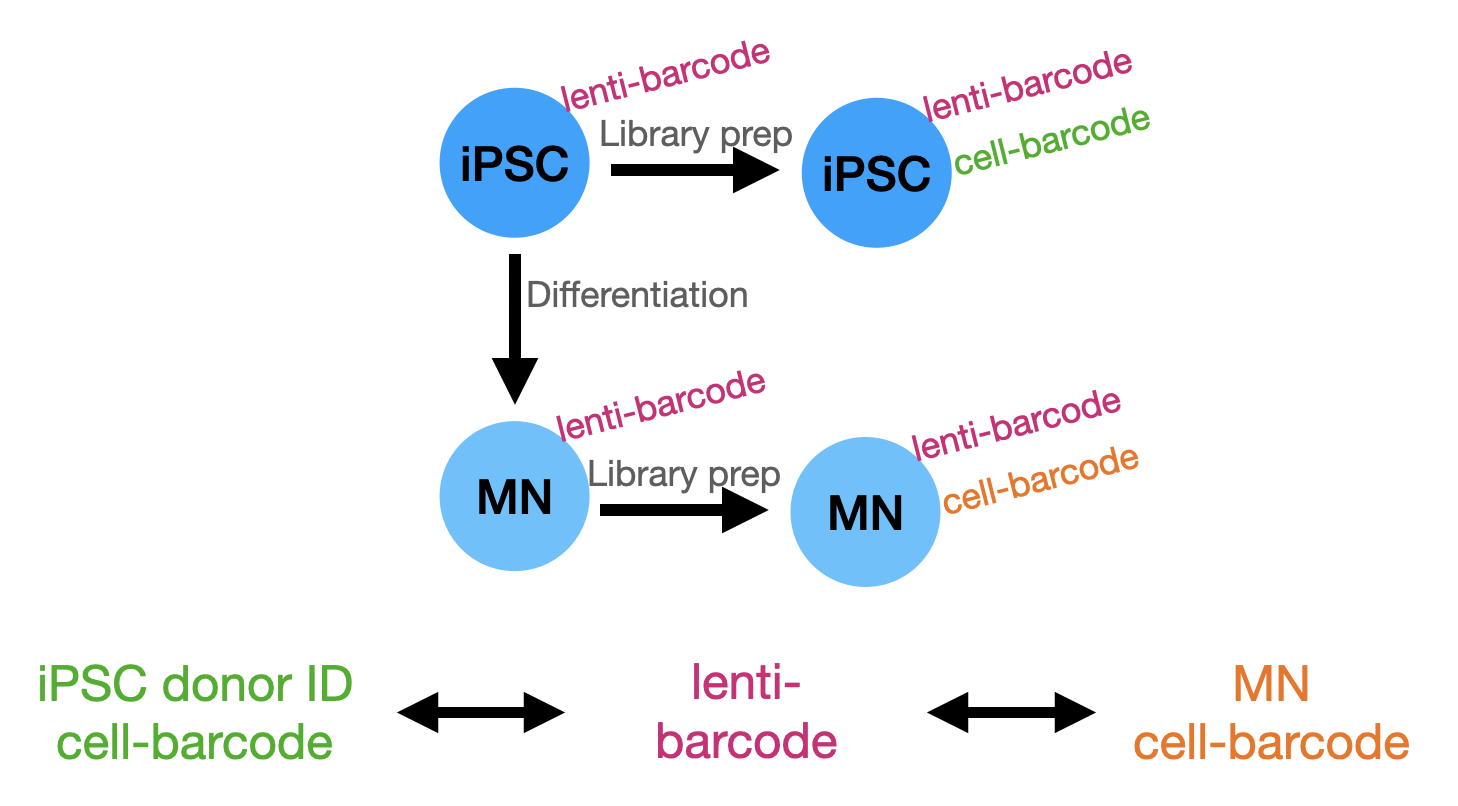
Overview of strategy
Comparing lenti-barcode map to called cells
First assessing how often called cells (by cellranger for now, but eventually using the ensemble cell calling approach) were also assigned lenti barcodes for Capture 4 and 5 separately. Action item: rerun this with the iPSC cells after increasing the cell threshold on cellbender - a quick test showed that if I included all barcodes called as cells by 2+ methods for the iPSCs, then about 90% of lenti barcoded cell barcodes were a called iPSC!
c4_lenti_key <- read.csv(paste0(lenti_dir, "Capture4-Lenti_cellbarcodes.tsv"),
sep = "\t")
c5_lenti_key <- read.csv(paste0(lenti_dir, "Capture5-Lenti_cellbarcodes.tsv"),
sep = "\t")
c4_lenti_key <- c4_lenti_key %>% separate_rows(cell_barcodes, sep=";", convert = TRUE) %>%
mutate(cell_barcodes = paste0(cell_barcodes, "-1"))
c5_lenti_key <- c5_lenti_key %>% separate_rows(cell_barcodes, sep=";", convert = TRUE) %>%
mutate(cell_barcodes = paste0(cell_barcodes, "-1"))
c4_cellbarcodes <- scan(paste0(mn_dir, "01_cellcalling-merged/Capture4-GEX/barcodes.txt"), what="character")
c5_cellbarcodes <- scan(paste0(ipsc_dir, "01_cellcalling-merged/Capture5-GEX/barcodes.txt"), what="character")
c4_ovlp <- table(c4_lenti_key$cell_barcodes %in% c4_cellbarcodes)
c5_ovlp <- table(c5_lenti_key$cell_barcodes %in% c5_cellbarcodes)
message("lenti barcodes that belong to a called MN cell: ", c4_ovlp[["TRUE"]], " (", round(c4_ovlp[["TRUE"]]/nrow(c4_lenti_key)*100, 1), "%)")lenti barcodes that belong to a called MN cell: 1695 (20.7%)message("lenti barcodes that belong to a called iPSC: ", c5_ovlp[["TRUE"]], " (", round(c5_ovlp[["TRUE"]]/nrow(c4_lenti_key)*100, 1), "%)")lenti barcodes that belong to a called iPSC: 2031 (24.9%)However, this is still a relatively small proportion of called cells in both captures:
c4_ovlp2 <- table(c4_cellbarcodes %in% c4_lenti_key$cell_barcodes)
c5_ovlp2 <- table(c5_cellbarcodes %in% c5_lenti_key$cell_barcodes)
message("Called MNs with a lenti barcode: ", c4_ovlp2[["TRUE"]], " (", round(c4_ovlp2[["TRUE"]]/length(c4_cellbarcodes)*100, 1), "%)")Called MNs with a lenti barcode: 1695 (8.3%)message("Called iPSCs with a lenti barcode: ", c5_ovlp2[["TRUE"]], " (", round(c5_ovlp2[["TRUE"]]/length(c5_cellbarcodes)*100, 1), "%)")Called iPSCs with a lenti barcode: 2031 (10.7%)Comparison with donor calls from vireo
Multiple cells may have the same lenti-barcode, but all of these cells should be from the same donor.
iPSCs (Capture 5)
First, merge the lenti-barcode information with the donor calls from vireo for the iPSCs. How many donors are assigned to each lenti-barcode?
ipsc_donors <- read.csv(paste0(ipsc_dir, "03_vireo-missing-some-cells/03_vireo/Capture5-GEX/donor_ids.tsv"), sep="\t")
ipsc_donors$cell_barcodes <- ipsc_donors$cell
sort(table(ipsc_donors$donor_id))
130 141 191 199 75 82 87
1 1 1 1 1 1 1
98 T232 W103 W221 105 110 118
1 1 1 1 2 2 2
121 202 239 240 74 797 99
2 2 2 2 2 2 2
donor138 donor141 W104 W134 W162 113 131
2 2 2 2 2 3 3
217 donor137 donor145 90 W222 208 220
3 3 3 4 4 5 5
238 93 donor140 T234 W001 194 donor144
5 5 5 5 5 6 6
donor147 W263 124 donor143 donor142 196 76
6 6 8 8 9 10 10
donor139 donor146 123 231 221 donor149 W112
10 10 11 11 12 12 12
donor148 227 donor136 W014 107 92 237
13 14 14 14 16 19 24
127 214 102 192 104 210 190
25 27 28 39 47 48 50
218 W005 189 128 187 207 122
52 63 70 71 78 83 90
W187 100 W132 88 198 154 119
91 101 147 155 286 290 307
129 197 doublet unassigned
1627 3632 4530 6702 ipsc_donors <- ipsc_donors %>% left_join(c5_lenti_key[, c("lenti_bc", "cell_barcodes")],
by="cell_barcodes")
ipsc_donors_lenti <- na.omit(ipsc_donors)
message(length(unique(ipsc_donors_lenti$lenti_bc)), " lenti barcodes map to ",
length(unique(ipsc_donors_lenti$cell_barcodes)), " iPSCs") 197 lenti barcodes map to 2031 iPSCsipsc_donors_lenti <- subset(ipsc_donors_lenti, ! donor_id %in% c("unassigned", "doublet") )
message(length(unique(ipsc_donors_lenti$lenti_bc)), " lenti barcodes map to ",
length(unique(ipsc_donors_lenti$cell_barcodes)), " iPSCs with donor IDs from vireo.") 147 lenti barcodes map to 1134 iPSCs with donor IDs from vireo.summary_ipsc_donors_lenti <- ipsc_donors_lenti %>%
group_by(donor_id, lenti_bc) %>% tally() %>% group_by(lenti_bc) %>%
tally()
message("Number of lenti barcodes mapping to n donor IDs:")Number of lenti barcodes mapping to n donor IDs:table(summary_ipsc_donors_lenti$n)
1 2 18
137 9 1 For lenti barcodes associated with more than one donor, what proportion (text=count) of cells are assigned to different donors?
inspect_bcs <- subset(summary_ipsc_donors_lenti, n > 1)$lenti_bc
ipsc_donors_lenti %>%
filter(lenti_bc %in% inspect_bcs) %>%
ggplot(aes(fill=donor_id, x=lenti_bc)) +
geom_bar(position="fill") +
scale_y_continuous(labels = scales::percent, breaks = c(0, 0.2, 0.4, 0.6, 0.8, 1)) +
theme_cowplot() +
geom_text(stat="count",aes(label=ifelse((..count..)>0, ..count.., "")),
position = position_fill(vjust=0.5)) +
coord_flip()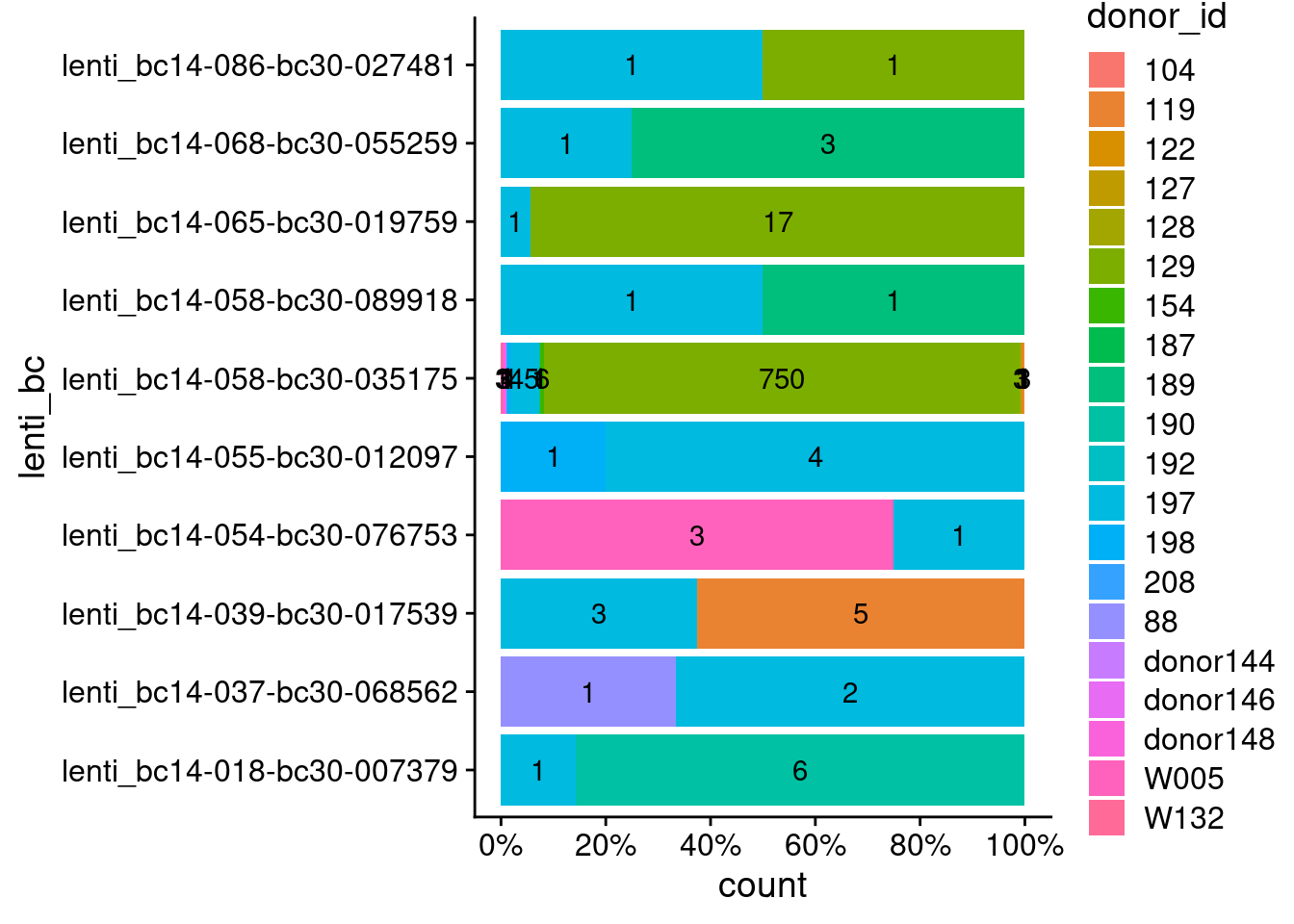
Number of cell barcodes (x-axis) associated with different donors (fill color) for each lenti barcode (y-axis).
| Version | Author | Date |
|---|---|---|
| a1fa048 | cazodi | 2022-03-01 |
Ignoring these lenti barcodes with ambiguous donor IDs, can we use the other lenti barcodes to assign donorIDs to more cells? Cells called as doublets by vireo should remain doublets, but for those that were unassigned by vireo, how many could we assign using a lenti barcode and how often do the lenti assignments match the best singlet estimated by vireo? This may give us a sense of how well we will be able to do this in MN cells…
good_lenti_barcodes <- subset(summary_ipsc_donors_lenti, n == 1)$lenti_bc
lenti_donor_key <- subset(ipsc_donors_lenti, lenti_bc %in% good_lenti_barcodes) %>%
distinct(lenti_bc, donor_id)
ipsc_donors <- ipsc_donors %>% left_join(lenti_donor_key, by = "lenti_bc", suffix=c("", ".lenti"))
ipsc_donors_no_vireoID <- subset(ipsc_donors, donor_id %in% c("unassigned"))
x <- nrow(ipsc_donors_no_vireoID)
ipsc_donors_no_vireoID <- subset(ipsc_donors_no_vireoID, !is.na(donor_id.lenti))
x2 <- nrow(ipsc_donors_no_vireoID)
res <- table(ipsc_donors_no_vireoID$best_singlet == ipsc_donors_no_vireoID$donor_id.lenti)
message("Of ", x, " unassigned cells, ", x2, " have are associated with an unambiguous lenti barcodes and of those, ", res[["TRUE"]], " (", round(res[["TRUE"]]/x2*100, 1),"%) are the same as the best singlet estimated by vireo.")Of 6702 unassigned cells, 76 have are associated with an unambiguous lenti barcodes and of those, 48 (63.2%) are the same as the best singlet estimated by vireo.Inspecting donor 197
Does donor 197 have a proportionately large number of lenti barcodes? Or did a very large number of cells come from a single (lenti barcoded) progenitor?
ipsc_donors %>% group_by(donor_id) %>%
filter(!donor_id %in% c("doublet", "unassigned")) %>%
summarise(n_cells = n(),
n_lentiBCs = length(unique(lenti_bc))) %>%
ggscatter(x="n_lentiBCs", y="n_cells", label = "donor_id", repel = TRUE, add = "reg.line")`geom_smooth()` using formula 'y ~ x'Warning: ggrepel: 81 unlabeled data points (too many overlaps). Consider
increasing max.overlaps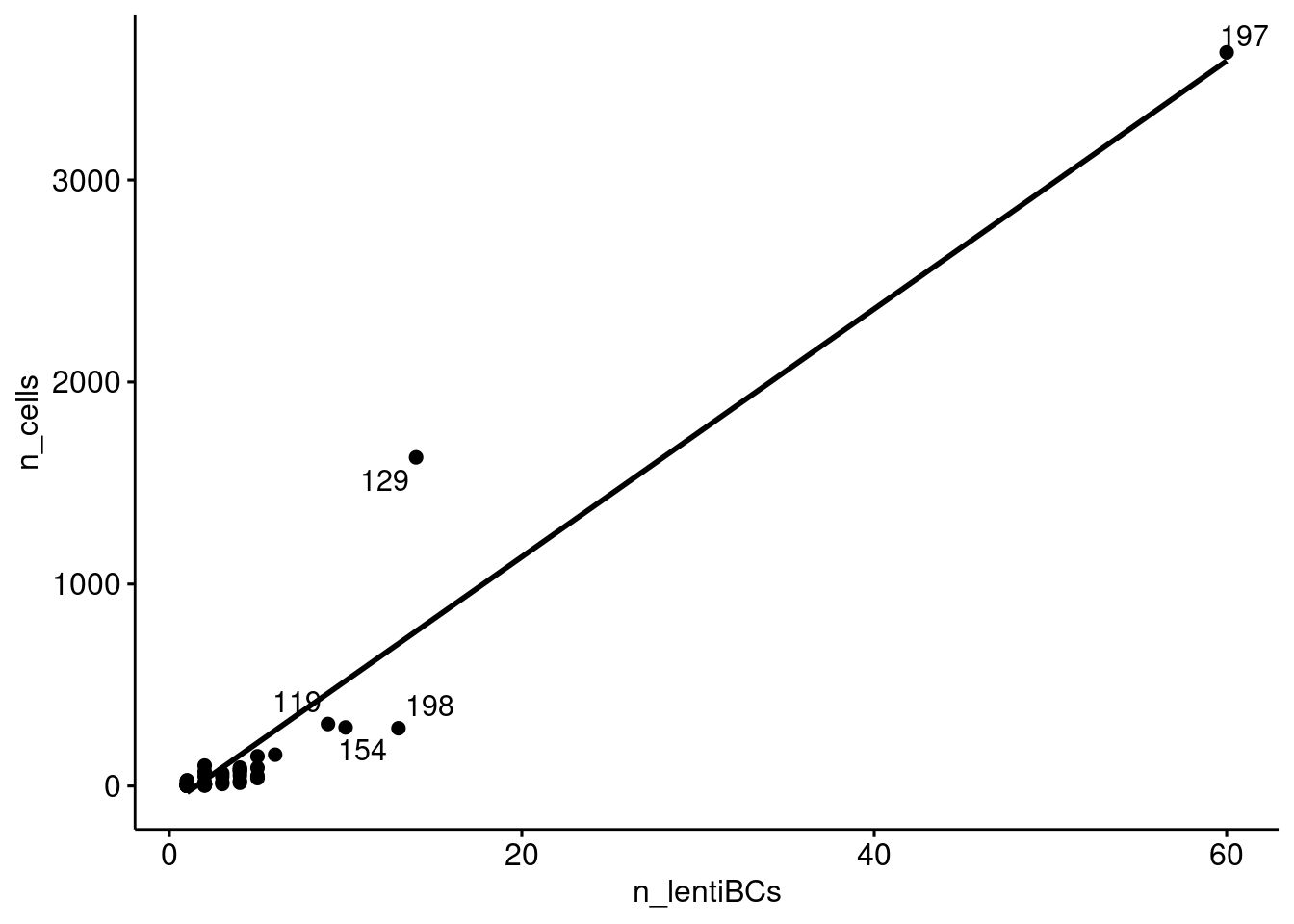
Relationship between the number of cells assigned to a donor and the number of lenti barcodes assigned to a donor.
| Version | Author | Date |
|---|---|---|
| a1fa048 | cazodi | 2022-03-01 |
d197 <- ipsc_donors %>% filter(donor_id == "197")
d197_lbc_counts <- as.data.frame(table(d197$lenti_bc))
summary(d197_lbc_counts$Freq) Min. 1st Qu. Median Mean 3rd Qu. Max.
1.000 1.000 1.000 2.797 2.000 45.000 MN donor identification
Using the lenti barcodes with an unambiguous donorID from iPSCs, assign donor IDs to MNs.
c4_lenti_key_cells <- subset(c4_lenti_key, cell_barcodes %in% c4_cellbarcodes)
c4_donor_assignments <- c4_lenti_key_cells %>% left_join(lenti_donor_key, by="lenti_bc")
n_assigned <- nrow(na.omit(c4_donor_assignments))
message("Number of cells assigned: ", n_assigned, " (",
round(n_assigned/nrow(c4_donor_assignments)*100, 2), "% of MN cells)")Number of cells assigned: 471 (27.79% of MN cells)message("Number of cells assigned to each donor:")Number of cells assigned to each donor:sort(table(c4_donor_assignments$donor_id))
187 192 92 194 W221 W187 154 198 119 189 W132 129 88 197
1 1 1 2 2 3 8 10 14 17 17 28 39 328 message("Percent of cells assigned to each donor:")Percent of cells assigned to each donor:round(sort(table(c4_donor_assignments$donor_id))/n_assigned *100, 2)
187 192 92 194 W221 W187 154 198 119 189 W132 129 88
0.21 0.21 0.21 0.42 0.42 0.64 1.70 2.12 2.97 3.61 3.61 5.94 8.28
197
69.64 Consistent with iPSC analysis, 51.1% of MN cells that could be assigned a donor are from donor 197 (46.7% of iPSCs). However, donor 129 was also enriched in the iPSCs (20.9% of donor identified iPSCs), but is not in the MNs (4.8% of donor IDed MNs). This, in addition to the lenti barcode (“lenti_bc14-058-bc30-035175”) with 750 cells all from donor 129, suggests that the progenitor cell with that lenti barcode out competed other cells after being lenti barcoding and FACS sorting (i.e. during the paralle culture phase). While donor 197 cells seemed to have been more abundant earlier on in the experiment, perhaps from the start or maybe they took up more lenti barcodes and were then enriched after FACS sorting.
Comparing lenti donor IDs with vireo results for MNs
sessionInfo()R version 4.1.1 (2021-08-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Red Hat Enterprise Linux 8.5 (Ootpa)
Matrix products: default
BLAS/LAPACK: /usr/lib64/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] viridis_0.6.2 viridisLite_0.4.0 cowplot_1.1.1 ggpubr_0.4.0
[5] ggplot2_3.3.5 data.table_1.14.2 tidyr_1.1.4 dplyr_1.0.7
[9] argparse_2.1.3
loaded via a namespace (and not attached):
[1] ggrepel_0.9.1 Rcpp_1.0.7 lattice_0.20-45 assertthat_0.2.1
[5] rprojroot_2.0.2 digest_0.6.29 utf8_1.2.2 R6_2.5.1
[9] backports_1.4.1 evaluate_0.14 highr_0.9 pillar_1.6.4
[13] rlang_0.4.12 whisker_0.4 car_3.0-12 jquerylib_0.1.4
[17] Matrix_1.4-0 rmarkdown_2.11 labeling_0.4.2 splines_4.1.1
[21] stringr_1.4.0 munsell_0.5.0 broom_0.7.10 compiler_4.1.1
[25] httpuv_1.6.5 xfun_0.28 pkgconfig_2.0.3 mgcv_1.8-38
[29] htmltools_0.5.2 tidyselect_1.1.1 tibble_3.1.6 gridExtra_2.3
[33] workflowr_1.6.2 fansi_1.0.0 crayon_1.4.2 withr_2.4.3
[37] later_1.3.0 grid_4.1.1 nlme_3.1-153 jsonlite_1.7.2
[41] gtable_0.3.0 lifecycle_1.0.1 DBI_1.1.1 git2r_0.29.0
[45] magrittr_2.0.1 scales_1.1.1 stringi_1.7.6 carData_3.0-4
[49] farver_2.1.0 ggsignif_0.6.3 fs_1.5.2 promises_1.2.0.1
[53] bslib_0.3.1 ellipsis_0.3.2 generics_0.1.1 vctrs_0.3.8
[57] tools_4.1.1 glue_1.6.0 purrr_0.3.4 abind_1.4-5
[61] fastmap_1.1.0 yaml_2.2.1 colorspace_2.0-2 rstatix_0.7.0
[65] knitr_1.36 sass_0.4.0