Lenti barcode analysis- just capture 5
Christina Azodi
2022-03-08
Last updated: 2022-03-08
Checks: 5 2
Knit directory: BAUH_2020_MND-single-cell/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/01_fasta/ | ../output/pilot3_Lenti/01_fasta |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/02_blast/ | ../output/pilot3_Lenti/02_blast |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/03_keys/ | ../output/pilot3_Lenti/03_keys |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/01_cellcalling-merged/Capture5-GEX/barcodes.txt | ../output/pilot3.0_iPSC/01_cellcalling-merged/Capture5-GEX/barcodes.txt |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/pilot3_lenti_barcodes_capture5_poolA_D42pass.txt | ../data/pilot3_lenti_barcodes_capture5_poolA_D42pass.txt |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/Cellecta-SEQ-CloneTracker-XP_14bp_barcodes.txt | ../data/Cellecta-SEQ-CloneTracker-XP_14bp_barcodes.txt |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/Cellecta-SEQ-CloneTracker-XP_30bp_barcodes.txt | ../data/Cellecta-SEQ-CloneTracker-XP_30bp_barcodes.txt |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo/Capture5-GEX/donor_ids.tsv | ../output/pilot3.0_iPSC/03_vireo/Capture5-GEX/donor_ids.tsv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 77c6459. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .cache/
Ignored: .config/
Ignored: .snakemake/
Ignored: BAUH_2020_MND-single-cell.Rproj
Ignored: GRCh38_turboGFP-RFP_reference/
Ignored: Homo_sapiens.GRCh38.turboGFP/
Ignored: Rplots.pdf
Ignored: data/1-s2.0-S0002929720300781-main.pdf
Ignored: data/2103.11251.pdf
Ignored: data/3M-february-2018.txt
Ignored: data/737K-august-2016.txt
Ignored: data/Cellecta-SEQ-CloneTracker-XP_14bp_barcodes.txt
Ignored: data/Cellecta-SEQ-CloneTracker-XP_30bp_barcodes.txt
Ignored: data/STAR_index/
Ignored: data/STAR_output/
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.log
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.vcf.gz
Ignored: data/genome1k.chr22.log
Ignored: data/genome1k.chr22.recode.vcf
Ignored: data/pilot3_aggr-experiments.csv
Ignored: data/pilot3_donors.txt
Ignored: data/pilot3_lenti_barcodes_capture5_poolA_D42pass.txt
Ignored: data/s41588-018-0268-8.pdf
Ignored: data/tr2g_hs.tsv
Ignored: logs/
Ignored: output/2021-04-27_pilot2_nCells-per-donor.pdf
Ignored: output/2021-08-03_pilot2_nCells-per-donor.pdf
Ignored: output/CB-scRNAv31-GEX-lib01_QC_metadata.txt
Ignored: output/CB-scRNAv31-GEX-lib02_QC_metadata.txt
Ignored: output/pilot1_starsoloED/
Ignored: output/pilot2.1_gex/
Ignored: output/pilot2_HTO-2/
Ignored: output/pilot2_HTO/
Ignored: output/pilot2_gex_MAF01-152/
Ignored: output/pilot2_gex_MAF01/
Ignored: output/pilot2_gex_starsolo/
Ignored: output/pilot2_gex_starsoloED/
Ignored: output/pilot2_gex_starsoloED_GFP/
Ignored: output/pilot2_testing/
Ignored: output/pilot3.0/
Ignored: output/pilot3.0_MN/
Ignored: output/pilot3.0_captures-separate/
Ignored: output/pilot3.0_iPSC/
Ignored: output/pilot3_Lenti/
Ignored: references/Homo_sapiens.GRCh38.turboGFP.bed
Ignored: references/Homo_sapiens.GRCh38.turboGFP.fa
Ignored: references/Homo_sapiens.GRCh38.turboGFP.fa.fai
Ignored: references/Homo_sapiens.GRCh38.turboGFP.filtered.gtf
Ignored: references/Homo_sapiens.GRCh38.turboGFP.gtf
Ignored: references/SAindex/
Ignored: references/geno_test.vcf.gz
Ignored: references/pilot3/
Ignored: references/test
Ignored: references/turboGFP.fa
Ignored: references/turboGFP.gtf
Ignored: references/turboRFP.fa
Ignored: testupset_plot.pdf
Ignored: testvenn_diagram.pdf
Ignored: workflow/rules/
Untracked files:
Untracked: .nv/
Untracked: Capture5-GEX/
Untracked: __Capture5-GEX.mro
Untracked: analysis/2022-03-04_pilot3_LentiBarcodesCapture5.Rmd
Untracked: analysis/2022-03-04_pilot3_iPSC_donor_assignment.Rmd
Untracked: analysis/2022-03-06_pilot3_CellQC.Rmd
Untracked: analysis/figure/
Untracked: cellbender.dockerfile
Untracked: hwe1e-05_maf05_vcf_stats.txt
Untracked: hwe1e-05_vcf_stats.txt
Untracked: testbarcodes.txt
Untracked: testcombination_matrix.rds
Untracked: workflow/demuxlet_jobs/
Unstaged changes:
Modified: analysis/2022-02-24_pilot3_LentiBarcodes.Rmd
Modified: code/function_vireo.R
Modified: code/get_barcodes_to_use.R
Modified: config/config_pilot3.0_MN.yml
Modified: config/config_pilot3.0_iPSC.yml
Modified: workflow/Snakefile
Modified: workflow/rules_cellcalling.smk
Modified: workflow/rules_demultiplexing.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
suppressPackageStartupMessages({
library(argparse)
library(dplyr)
library(tidyr)
library(data.table)
library(ggpubr)
library(cowplot)
library(viridis)
library(stringr)
})
fasta_out <- "/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/01_fasta/"
blast_out <- "/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/02_blast/"
out <- paste0("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3_Lenti/03_keys/")
save <- TRUE
c5_cellbarcodes <- scan("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/01_cellcalling-merged/Capture5-GEX/barcodes.txt", what="character")
c5_cellbarcodes <- gsub("-1", "", c5_cellbarcodes)Goal
Generate a map between iPSC cell barcodes (Capture 5) and MN cell barcodes (Capture 4) by identifying the lenti barcode (14bp+30bp) shared by the two.
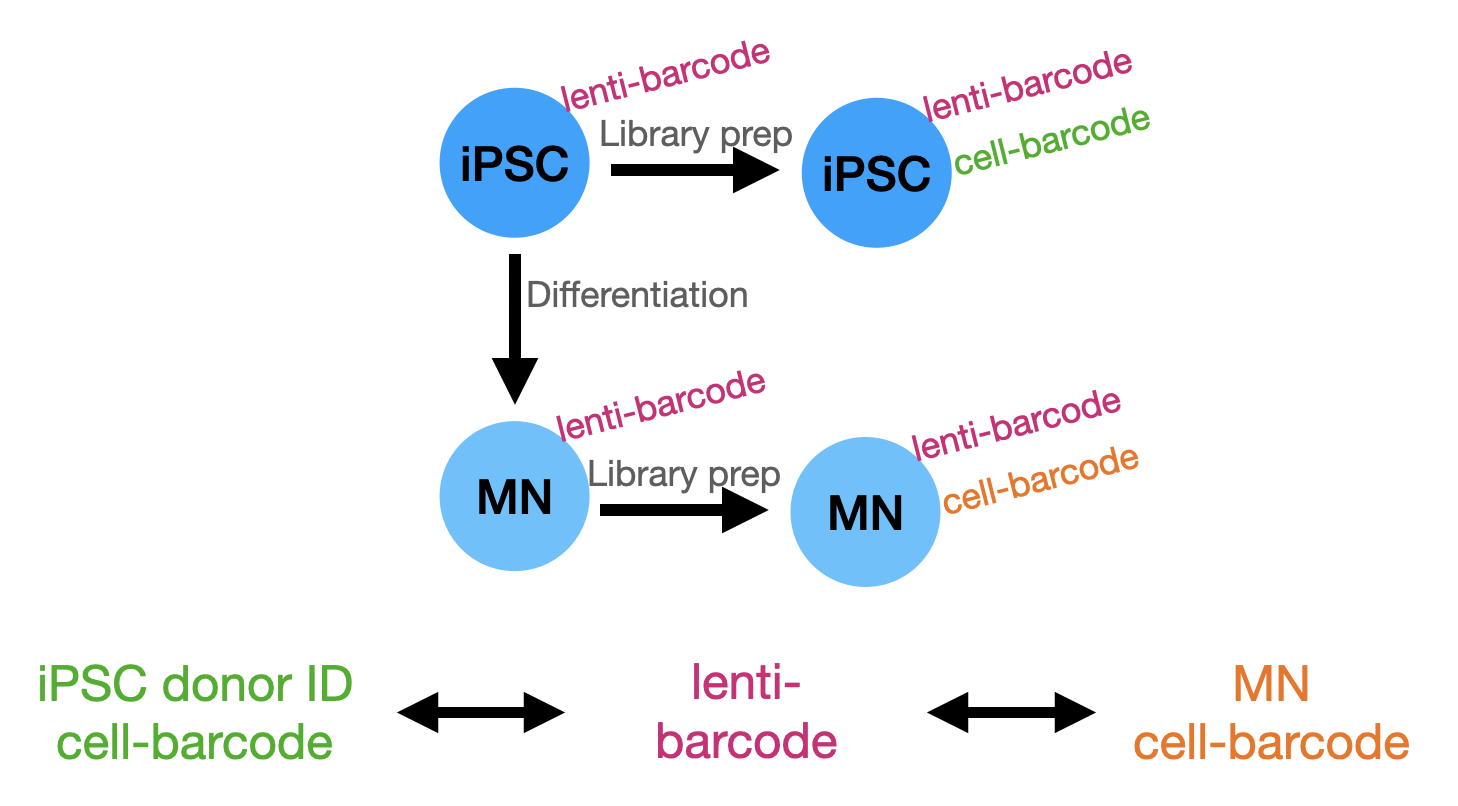
Overview of strategy
Blastn parsing
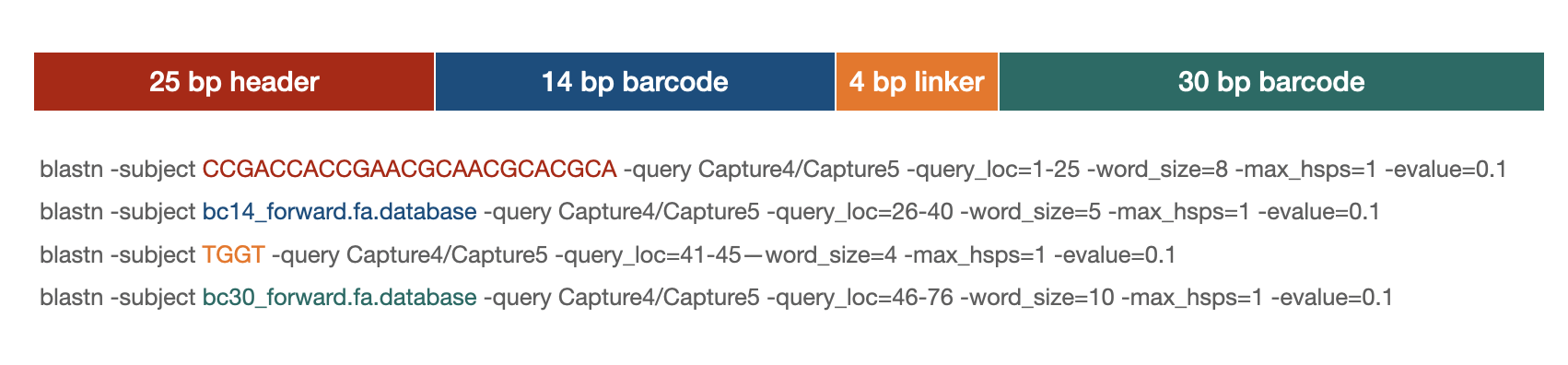
blastn strategy
Example scripts:
14 bp barcode: blastn -db output/pilot3_Lenti/00_sequences/bc14_forward.fa -query output/pilot3_Lenti/01_fasta/Capture5-Lenti.fa -query_loc 26-40 -word_size 5 -max_hsps 1 -evalue 0.1 -num_threads 4 -outfmt 6 -out output/pilot3_Lenti/02_blast/Capture5-Lenti_bc14_blast.out
30 bp barcode: blastn -db output/pilot3_Lenti/00_sequences/bc30_forward.fa -query output/pilot3_Lenti/01_fasta/Capture5-Lenti.fa -query_loc 46-76 -word_size 10 -max_hsps 1 -evalue 0.1 -max_target_seqs 1 -outfmt 6 -out output/pilot3_Lenti/02_blast/Capture5-Lenti_bc30_blast.out
Read in blastn hits and remove ambiguous matches by first keeping the longest hit for each read and if two lenti barcodes have equally long hits, removing the read entirely. Number of reads with lenti barcode matches:
load_parse_blastn <- function(path) {
df <- fread(path, header=FALSE, select = c(1:6))
names(df) <- c("qseqid", "sseqid", "pident", "length", "mismatch", "gapopen")
# If >1 hit for a lenti barcode, keep the one(s) with the longer overlap
df <- df[df[, .I[length == max(length)], by=qseqid]$V1]
# If >1 hit for a lenti barcode with equal lengths, remove them both.
df <- df[!(duplicated(df$qseqid) | duplicated(df$qseqid, fromLast = TRUE)), ]
return(df)
}
## Load and process blastn results
c5_bc14 <- load_parse_blastn(paste0(blast_out, "Capture5-Lenti_bc14_blast.out"))
c5_bc30 <- load_parse_blastn(paste0(blast_out, "Capture5-Lenti_bc30_blast.out"))
# Merge
c5_both <- c5_bc14 %>% inner_join(c5_bc30, by = "qseqid", suffix = c(".bc14", ".bc30"))
c5_both_safe <- c5_both
lenti_stats <- as.data.frame(list(capture=c( "Capture 5"),
raw=c( 46041484),
bc14=c(nrow(c5_bc14)),
bc30=c(nrow(c5_bc30)),
bc14_and_30=c(nrow(c5_both))))
#rm(c5_bc14, c5_bc30)Filtering by lenti-barcode quality/inclustion
Because we are relying primarily on the lenti barcodes for QC (as the header 25 bp and linker 4 bp regions are low quality), we want strict matching criteria. Thus we will remove any reads with lenti barcode mismatches or with length < 14/28 for the 14bp and 30 bp barcode, respectively.
# Add lenti barcode IDs
c5_both$lenti_bc <- paste0("lenti_", c5_both$sseqid.bc14, "-", c5_both$sseqid.bc30)
# Filtering by minimum length
c5_both <- subset(c5_both, c5_both$length.bc14 == 14 & c5_both$length.bc30 >= 28)
# Filtering by maximum mismatches
c5_both <- subset(c5_both, c5_both$mismatch.bc14 == 0 & c5_both$mismatch.bc30 == 0)
lenti_stats$strict_filter <- c(nrow(c5_both))
t(lenti_stats %>% mutate_at(vars(3:6), list(percent_raw=~./raw*100)) %>%
mutate(across(where(is.numeric), round, 1))) [,1]
capture "Capture 5"
raw "46041484"
bc14 "17260005"
bc30 "12795040"
bc14_and_30 "12544555"
strict_filter "11403761"
bc14_percent_raw "37.5"
bc30_percent_raw "27.8"
bc14_and_30_percent_raw "27.2"
strict_filter_percent_raw "24.8" Further, we known which lenti barcodes were used to tag the iPSCs, so we can remove any that are assigned a lenti barcode not used.
known_bc <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/pilot3_lenti_barcodes_capture5_poolA_D42pass.txt",
sep="\t", header=TRUE)
names(known_bc) <- c("BC14_rc", "BC30_rc")
bc14_names <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/Cellecta-SEQ-CloneTracker-XP_14bp_barcodes.txt",
sep="\t", header=TRUE)
bc30_names <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/Cellecta-SEQ-CloneTracker-XP_30bp_barcodes.txt",
sep="\t", header=TRUE)
known_bc <- known_bc %>% left_join(bc14_names, by="BC14_rc")
known_bc <- known_bc %>% left_join(bc30_names, by="BC30_rc")
known_bc$lenti_bc <-paste0("lenti_", known_bc$BC14_ID, "-", known_bc$BC30_ID)
known_bc$bc_number <- seq(1, nrow(known_bc))
known_ovlp <- table(c5_both$lenti_bc %in% known_bc$lenti_bc)
x <- nrow(c5_both)
c5_both <- subset(c5_both, lenti_bc %in% known_bc$lenti_bc)
message("C5 QCed lenti barcoded reads that are known lenti barcode combinations: ", nrow(c5_both), " (", round(nrow(c5_both) / x*100, 2), "%)")C5 QCed lenti barcoded reads that are known lenti barcode combinations: 11219678 (98.39%)Merge with cellbarcodes from the R1 reads
The R1 reads are all exactly 28 bp and contain the 16 bp cell barcode followed by a 12 bp UMI.
parse_merge_R1_cellbarcodes <- function(path, lenti) {
cbc <- fread(path, header=FALSE, sep="\t")
summary(nchar(cbc$cell_barcode))
names(cbc) <- c("qseqid", "R1")
cbc$qseqid <- gsub(">", "", gsub(" .*", "", cbc$qseqid))
cbc <- separate(cbc, R1, into = c("cell_barcode", "umi"), sep = 16)
lenti <- lenti %>% left_join(cbc, by = "qseqid")
rm(cbc)
return(lenti)
}
c5_both <- parse_merge_R1_cellbarcodes(paste0(fasta_out, "Capture5-Lenti_R1.tsv"), c5_both)
c5_both$cb_isCell <- c5_both$cell_barcode %in% c5_cellbarcodes
message("True cell barcodes remaining: ",
nrow(unique(c5_both[c5_both$cb_isCell==TRUE, "cell_barcode"])))True cell barcodes remaining: 34380if(save){ write.table(c5_both, paste0(out, "Capture5-Lenti_merged_barcodes.tsv"),
sep = "\t", quote=FALSE, row.names = FALSE)}Find unambiguous cell-to-lenti barcode pairs
Ideally, each cell should have been tagged with one lenti barcode, however there was nothing stopping more than one barcode being assigned to each cell.
c5_umis <- c5_both %>% filter(cb_isCell == TRUE) %>%
distinct(cell_barcode, lenti_bc, umi) %>%
group_by(cell_barcode, lenti_bc) %>%
tally(name = "umis") %>% group_by(cell_barcode) %>%
mutate(percent_umis_with_lentiBC = umis/sum(umis),
total_umis = sum(umis),
cb_isCell = cell_barcode %in% c5_cellbarcodes)
c5_umis %>% group_by(cell_barcode) %>% tally(name="n_lenti") %>%
ggplot(aes(x=n_lenti)) + geom_histogram(binwidth = 1) + theme_cowplot() 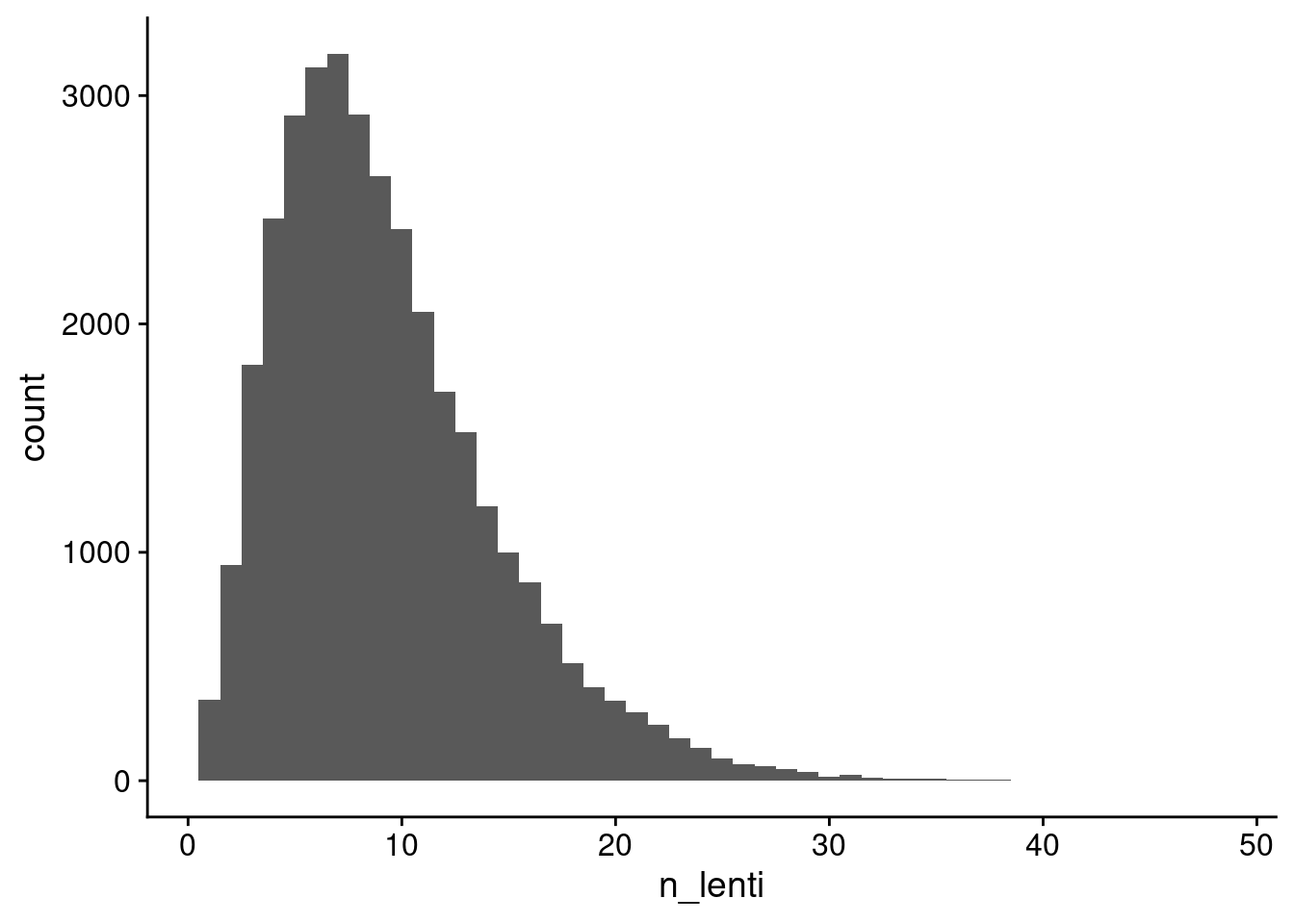
Distribution of the number of lenti-barcodes per cell barcode (removing cell barcodes not called as cells by the ensemble cell calling approach).
c5_umis %>% group_by(cell_barcode) %>%
filter(percent_umis_with_lentiBC == max(percent_umis_with_lentiBC)) %>%
ggplot(aes(x=percent_umis_with_lentiBC, y = total_umis)) +
geom_hex() + theme_cowplot() 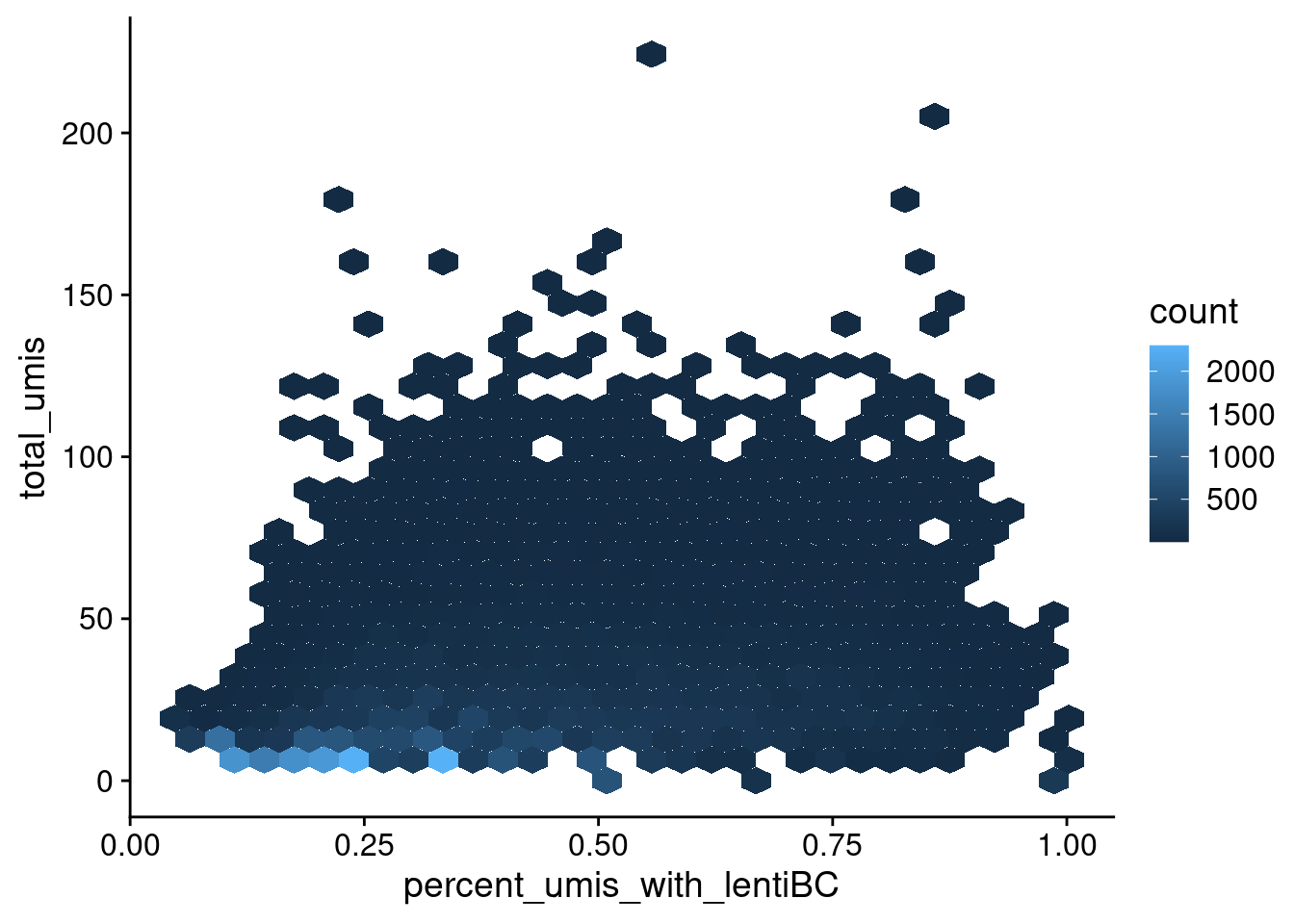
Relationship between total umis per cell barcode and the percent of umis with the most abundant lenti barcode for that cell.
perfect <- subset(c5_umis, percent_umis_with_lentiBC == 1)
message("Percent of cells with all UMIs mapping to the same lenti-barcode: ",
round(nrow(perfect) / length(unique(c5_umis$cell_barcode))*100, 2), "%")Percent of cells with all UMIs mapping to the same lenti-barcode: 1.03%Singular cell-lenti-barcode pairs
donors <- read.table("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo/Capture5-GEX/donor_ids.tsv", header=TRUE)
donors$cell_barcode <- gsub("-1", "", donors$cell)
perfect <- perfect %>%
left_join(donors[, c("cell_barcode", "donor_id", "best_singlet")],
by="cell_barcode")
message("Single lenti barcodes for ", nrow(perfect), " cells with ",
length(unique(perfect$lenti_bc)), " unique lenti barcodes")Single lenti barcodes for 355 cells with 127 unique lenti barcodesn_donors_per_lenti <- perfect %>%
filter(!donor_id %in% c("unassigned", "doublet")) %>%
group_by(lenti_bc, donor_id) %>% tally() %>%
group_by(lenti_bc) %>% tally()
message("Number of lenti barcodes with n donors assigned: ")Number of lenti barcodes with n donors assigned: table(n_donors_per_lenti$n)
1 2 4 21
75 13 1 1 Well supported single-lenti-cell barcode matches
At least 5 umis for the cell barcode with 80% of those umis supporting one barcode
supported <- subset(c5_umis, total_umis >= 5 &
percent_umis_with_lentiBC >= 0.8) %>%
left_join(donors[, c("cell_barcode", "donor_id", "best_singlet")],
by="cell_barcode")
message("Well supported lenti barcodes for ", nrow(supported), " cells with ",
length(unique(supported$lenti_bc)), " unique lenti barcodes")Well supported lenti barcodes for 1610 cells with 194 unique lenti barcodesn_donors_per_lenti <- supported %>%
filter(!donor_id %in% c("unassigned", "doublet")) %>%
group_by(lenti_bc, donor_id) %>% tally() %>%
group_by(lenti_bc) %>% tally()
message("Number of lenti barcodes with n donors assigned: ")Number of lenti barcodes with n donors assigned: table(n_donors_per_lenti$n)
1 2 22
149 2 1 if(save){write.table(perfect, paste0(out, "Capture5_lenti2donor_key.tsv"),
sep = "\t", quote=FALSE, row.names = FALSE)}Well supported multi-lenti to cell barcode matches
At least 5 umis support the lenti barcode-cell match and it makes up at least 20% of the support.
multi.supported <- subset(c5_umis, umis >= 5 &
percent_umis_with_lentiBC >= 0.2) %>%
arrange(lenti_bc) %>% group_by(cell_barcode) %>%
summarise(joint_lenti = toString(lenti_bc)) %>%
ungroup() %>% left_join(donors[, c("cell_barcode", "donor_id", "best_singlet")],
by="cell_barcode")
message("Well supported lenti barcodes for ", nrow(multi.supported), " cells with ",
length(unique(multi.supported$joint_lenti)), " unique lenti barcode combos")Well supported lenti barcodes for 22574 cells with 2895 unique lenti barcode combosn_donors_per_lenti <- multi.supported %>%
filter(!donor_id %in% c("unassigned", "doublet")) %>%
group_by(joint_lenti, donor_id) %>% tally() %>%
group_by(joint_lenti) %>% tally()
message("Number of lenti barcodes with n donors assigned: ")Number of lenti barcodes with n donors assigned: table(n_donors_per_lenti$n)
1 2 3 4 5 6 7 68
1197 166 53 9 12 3 1 1 sort(table(multi.supported$donor_id))
141 188 200 202 232 239 74
1 1 1 1 1 1 1
87 89 W103 110 121 130 138
1 1 1 2 2 2 2
75 797 93 W221 208 240 90
2 2 2 2 3 3 3
99 W134 W162 113 131 220 W001
3 3 3 4 4 4 4
W222 W263 118 donor148 T233 W104 105
4 4 5 5 5 5 6
231 124 donor139 donor144 donor138 238 196
6 7 7 7 8 10 11
donor140 donor143 T234 donor141 221 donor145 W112
11 11 11 12 13 13 13
76 donor142 donor146 123 donor147 194 donor137
14 15 15 16 17 19 19
donor136 W014 227 donor149 237 102 127
21 21 22 25 26 34 34
92 214 107 192 128 187 104
34 38 39 44 50 69 74
190 210 W005 W187 189 207 218
84 87 96 105 108 110 111
100 122 W132 88 198 154 119
115 123 188 201 355 394 437
129 197 doublet unassigned
2979 4075 5517 6609 if(save){write.table(multi.supported, paste0(out, "Capture5_lenti2donor_key.tsv"),
sep = "\t", quote=FALSE, row.names = FALSE)}
sessionInfo()R version 4.1.1 (2021-08-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Rocky Linux 8.5 (Green Obsidian)
Matrix products: default
BLAS/LAPACK: /usr/lib64/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] stringr_1.4.0 viridis_0.6.2 viridisLite_0.4.0 cowplot_1.1.1
[5] ggpubr_0.4.0 ggplot2_3.3.5 data.table_1.14.2 tidyr_1.1.4
[9] dplyr_1.0.7 argparse_2.1.3
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.28 bslib_0.3.1 purrr_0.3.4
[5] lattice_0.20-45 carData_3.0-5 colorspace_2.0-3 vctrs_0.3.8
[9] generics_0.1.2 htmltools_0.5.2 yaml_2.2.1 utf8_1.2.2
[13] rlang_0.4.12 hexbin_1.28.2 jquerylib_0.1.4 later_1.3.0
[17] pillar_1.6.4 glue_1.6.0 withr_2.4.3 DBI_1.1.2
[21] lifecycle_1.0.1 munsell_0.5.0 ggsignif_0.6.3 gtable_0.3.0
[25] workflowr_1.6.2 evaluate_0.15 labeling_0.4.2 knitr_1.36
[29] fastmap_1.1.0 httpuv_1.6.5 fansi_1.0.0 highr_0.9
[33] broom_0.7.10 Rcpp_1.0.7 promises_1.2.0.1 scales_1.1.1
[37] backports_1.4.1 jsonlite_1.8.0 abind_1.4-5 farver_2.1.0
[41] fs_1.5.2 gridExtra_2.3 digest_0.6.29 stringi_1.7.6
[45] rstatix_0.7.0 rprojroot_2.0.2 grid_4.1.1 tools_4.1.1
[49] magrittr_2.0.2 sass_0.4.0 tibble_3.1.6 car_3.0-12
[53] crayon_1.5.0 pkgconfig_2.0.3 ellipsis_0.3.2 assertthat_0.2.1
[57] rmarkdown_2.11 R6_2.5.1 git2r_0.29.0 compiler_4.1.1