Pilot3: Donor identification of iPSCs
C.B. Azodi
2022-03-30
Last updated: 2022-03-30
Checks: 6 1
Knit directory: BAUH_2020_MND-single-cell/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/code/function_vireo.R | ../code/function_vireo.R |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/pilot3_donors.txt | ../data/pilot3_donors.txt |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/01_cellcalling-merged/Capture5-GEX/barcodes.txt | ../output/pilot3.0_iPSC/01_cellcalling-merged/Capture5-GEX/barcodes.txt |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo/Capture5-GEX/donor_ids.tsv | ../output/pilot3.0_iPSC/03_vireo/Capture5-GEX/donor_ids.tsv |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo-common/Capture5-GEX/donor_ids.tsv | ../output/pilot3.0_iPSC/03_vireo-common/Capture5-GEX/donor_ids.tsv |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo-LearnGT/Capture5-GEX/donor_ids.tsv | ../output/pilot3.0_iPSC/03_vireo-LearnGT/Capture5-GEX/donor_ids.tsv |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_demuxlet.single | ../output/pilot3.0_iPSC/03_demuxlet.single |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo-nCount10/Capture5-GEX/GT_barcodes.tsv | ../output/pilot3.0_iPSC/03_vireo-nCount10/Capture5-GEX/GT_barcodes.tsv |
| /mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/ | ../output/pilot3.0_iPSC |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 99b9b47. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .cache/
Ignored: .config/
Ignored: .snakemake/
Ignored: BAUH_2020_MND-single-cell.Rproj
Ignored: GRCh38_turboGFP-RFP_reference/
Ignored: Homo_sapiens.GRCh38.turboGFP/
Ignored: Rplots.pdf
Ignored: data/1-s2.0-S0002929720300781-main.pdf
Ignored: data/2103.11251.pdf
Ignored: data/3M-february-2018.txt
Ignored: data/737K-august-2016.txt
Ignored: data/Cellecta-SEQ-CloneTracker-XP_14bp_barcodes.txt
Ignored: data/Cellecta-SEQ-CloneTracker-XP_30bp_barcodes.txt
Ignored: data/STAR_index/
Ignored: data/STAR_output/
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.log
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.recode.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz.csi
Ignored: data/genome1K.phase3.SNP_AF5e2.chr1toX.hg38.vcf.gz
Ignored: data/genome1k.chr22.log
Ignored: data/genome1k.chr22.recode.vcf
Ignored: data/pilot3_aggr-experiments.csv
Ignored: data/pilot3_donors.txt
Ignored: data/pilot3_lenti_barcodes_capture5_poolA_D42pass.txt
Ignored: data/s41588-018-0268-8.pdf
Ignored: data/tr2g_hs.tsv
Ignored: logs/
Ignored: output/2021-04-27_pilot2_nCells-per-donor.pdf
Ignored: output/2021-08-03_pilot2_nCells-per-donor.pdf
Ignored: output/CB-scRNAv31-GEX-lib01_QC_metadata.txt
Ignored: output/CB-scRNAv31-GEX-lib02_QC_metadata.txt
Ignored: output/pilot1_starsoloED/
Ignored: output/pilot2.1_gex/
Ignored: output/pilot2_HTO-2/
Ignored: output/pilot2_HTO/
Ignored: output/pilot2_gex_MAF01-152/
Ignored: output/pilot2_gex_MAF01/
Ignored: output/pilot2_gex_starsolo/
Ignored: output/pilot2_gex_starsoloED_GFP/
Ignored: output/pilot2_testing/
Ignored: output/pilot3.0_MN/
Ignored: output/pilot3.0_iPSC/
Ignored: output/pilot3_Lenti/
Ignored: references/Homo_sapiens.GRCh38.turboGFP.bed
Ignored: references/Homo_sapiens.GRCh38.turboGFP.fa
Ignored: references/Homo_sapiens.GRCh38.turboGFP.fa.fai
Ignored: references/Homo_sapiens.GRCh38.turboGFP.filtered.gtf
Ignored: references/Homo_sapiens.GRCh38.turboGFP.gtf
Ignored: references/Homo_sapiens.GRCh38.turboGFP_gene.gtf
Ignored: references/SAindex/
Ignored: references/geno_test.vcf.gz
Ignored: references/pilot3/
Ignored: references/test
Ignored: references/turboGFP.fa
Ignored: references/turboGFP.gtf
Ignored: references/turboRFP.fa
Ignored: workflow/rules/
Untracked files:
Untracked: .nv/
Untracked: Capture5-GEX/
Untracked: __Capture5-GEX.mro
Untracked: cellbender.dockerfile
Untracked: code/run_soupX_pilot3_capture3.R
Untracked: code/subsetting_mpileup.R
Untracked: hwe1e-05_maf05_vcf_stats.txt
Untracked: hwe1e-05_vcf_stats.txt
Unstaged changes:
Modified: .gitignore
Modified: code/run_soupX_2.R
Modified: config/config_pilot3.0_MN.yml
Modified: config/config_pilot3.0_iPSC.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/2022-03-04_pilot3_iPSC_donor_assignment.Rmd) and HTML (public/2022-03-04_pilot3_iPSC_donor_assignment.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d01f3b8 | cazodi | 2022-03-30 | testing snp filtering for vireo |
| html | d01f3b8 | cazodi | 2022-03-30 | testing snp filtering for vireo |
| Rmd | 2c95e3b | cazodi | 2022-03-09 | add stats to demulxiplexing results |
| html | 2c95e3b | cazodi | 2022-03-09 | add stats to demulxiplexing results |
| Rmd | c683543 | cazodi | 2022-03-09 | updated c5 lenti barcode analysis and donor assignment comparisons |
| html | c683543 | cazodi | 2022-03-09 | updated c5 lenti barcode analysis and donor assignment comparisons |
suppressPackageStartupMessages({
library(corrplot)
library(dplyr)
library(tidyverse)
library(RColorBrewer)
library(ComplexHeatmap)
library(data.table)
library(cowplot)
library(ggpubr)
})
source("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/code/function_vireo.R")
donors <- scan("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/data/pilot3_donors.txt", what="character")
d.cols <- data.frame(list(donor=c(donors, "doublet", "unassigned"),
type = c(rep("genotyped", length(donors)),
"doublet", "unassigned")))
cellbarcodes <- scan("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/01_cellcalling-merged/Capture5-GEX/barcodes.txt", what="character")Compare different approaches
- Vireo-common: common SNPs (genome1K.phase3.SNP_AF5e2.chr1toX.hg38.sort.vcf.gz) - 43,136 SNPs pass QC (minCount>100).
- Vireo: filtered genotype data - 56,171 SNPs pass QC (MAF>0.05, HWE>1e-05, pairwise_r<0.95, minCount>20)
- Vireo LearnGT: filtered genotype data - 56,171 SNPs pass QC (MAF>0.05, HWE>1e-05, pairwise_r<0.95, minCount>20)
- Demuxlet: filtered genotype data - 1,943,649 SNPs and 5,699,930 reads passing all filtering.
Snapshot of results:
vireo_best <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo/Capture5-GEX/donor_ids.tsv",
sep="\t")
vireo_common <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo-common/Capture5-GEX/donor_ids.tsv",
sep="\t")
vireo_learn <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo-LearnGT/Capture5-GEX/donor_ids.tsv",
sep="\t")
demux_all <- fread("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_demuxlet.single", sep="\t")
demux_top <- demux_all %>% group_by(BARCODE) %>%
filter(LLK1 == max(LLK1)) %>% select(cell = BARCODE, demuxlet = SM_ID)
donor_ids <- vireo_best[, c("cell", "donor_id")]
names(donor_ids) <- c("cell", "vireo")
donor_ids$vireo_common <- vireo_common$donor_id
donor_ids$vireo_learnGT <- vireo_learn$donor_id
donor_ids <- donor_ids %>% left_join(demux_top, by="cell")
head(donor_ids) cell vireo vireo_common vireo_learnGT demuxlet
1: AAACCCAAGCTCGAAG-1 197 donor67 197 197
2: AAACCCAAGGAAAGTG-1 129 donor61 129 129
3: AAACCCAAGGACTATA-1 154 donor95 154 154
4: AAACCCAAGGGTACAC-1 197 donor67 197 197
5: AAACCCAAGGTGCTAG-1 100 unassigned 100 100
6: AAACCCAAGGTTAAAC-1 197 donor67 197 197Vireo genotype guided vs. common SNPs
donor_ids_assigned <- subset(donor_ids, ! vireo %in% c("unassigned", "doublet")
& ! vireo_common %in% c("unassigned", "doublet"))
vireo_vireoC <- as.matrix(table(donor_ids_assigned$vireo,
donor_ids_assigned$vireo_common))
vireo_vireoC <- vireo_vireoC[order(rowSums(vireo_vireoC),decreasing=T),
order(colSums(vireo_vireoC),decreasing=T)]
Heatmap(log10(1+vireo_vireoC), name = "log10(nCells+1)",
cluster_rows = FALSE, cluster_columns = FALSE,
column_names_gp = gpar(fontsize = 5), row_names_gp = gpar(fontsize = 5),
col = colorRampPalette(brewer.pal(8, "YlOrRd"))(25))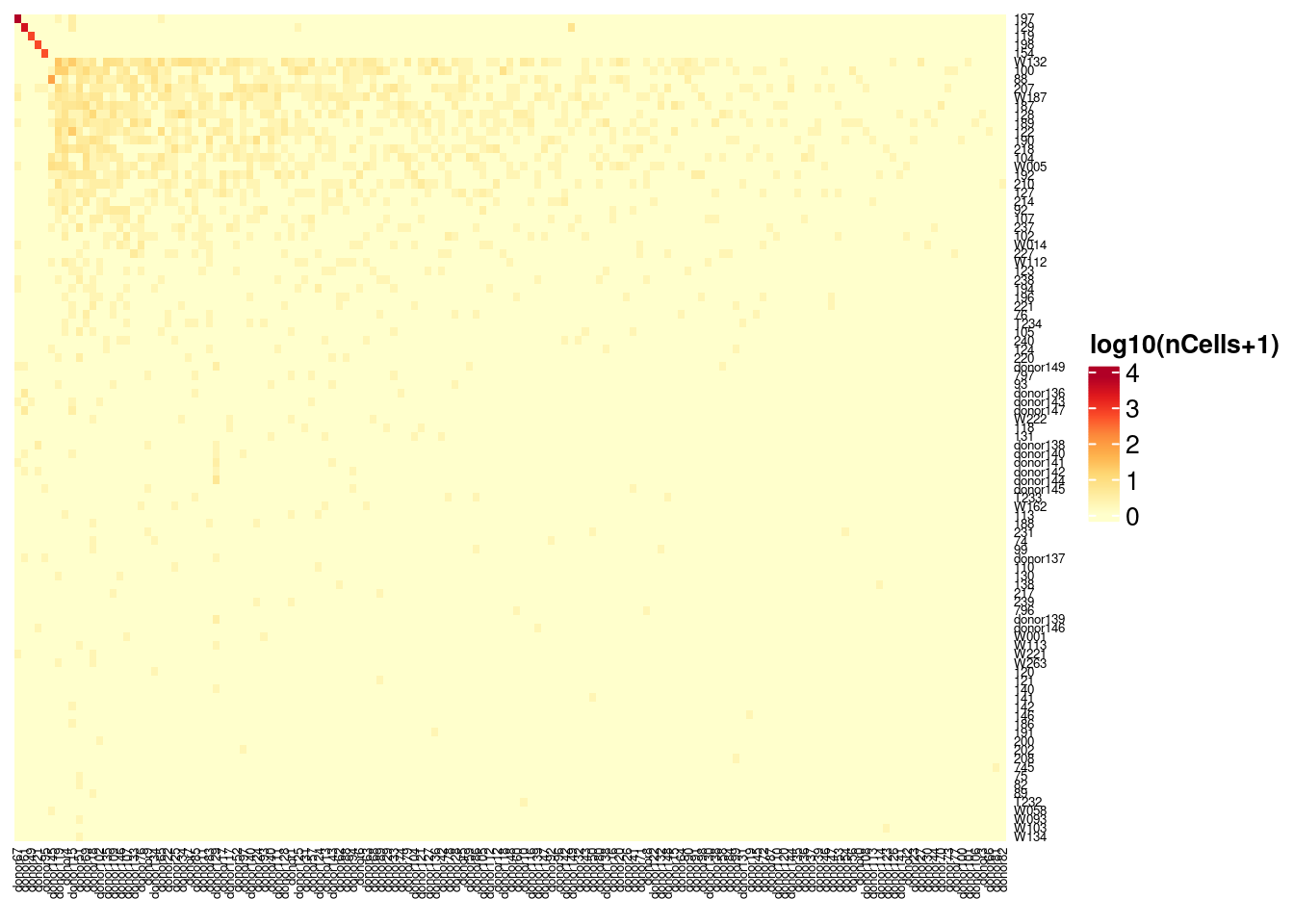
| Version | Author | Date |
|---|---|---|
| c683543 | cazodi | 2022-03-09 |
Zoom in on the 10 most abundant donors
Heatmap(vireo_vireoC[1:10,1:10], name = "nCells",
cluster_rows = FALSE, cluster_columns = FALSE,
column_names_gp = gpar(fontsize = 10), row_names_gp = gpar(fontsize = 10),
col = colorRampPalette(brewer.pal(8, "YlOrRd"))(25))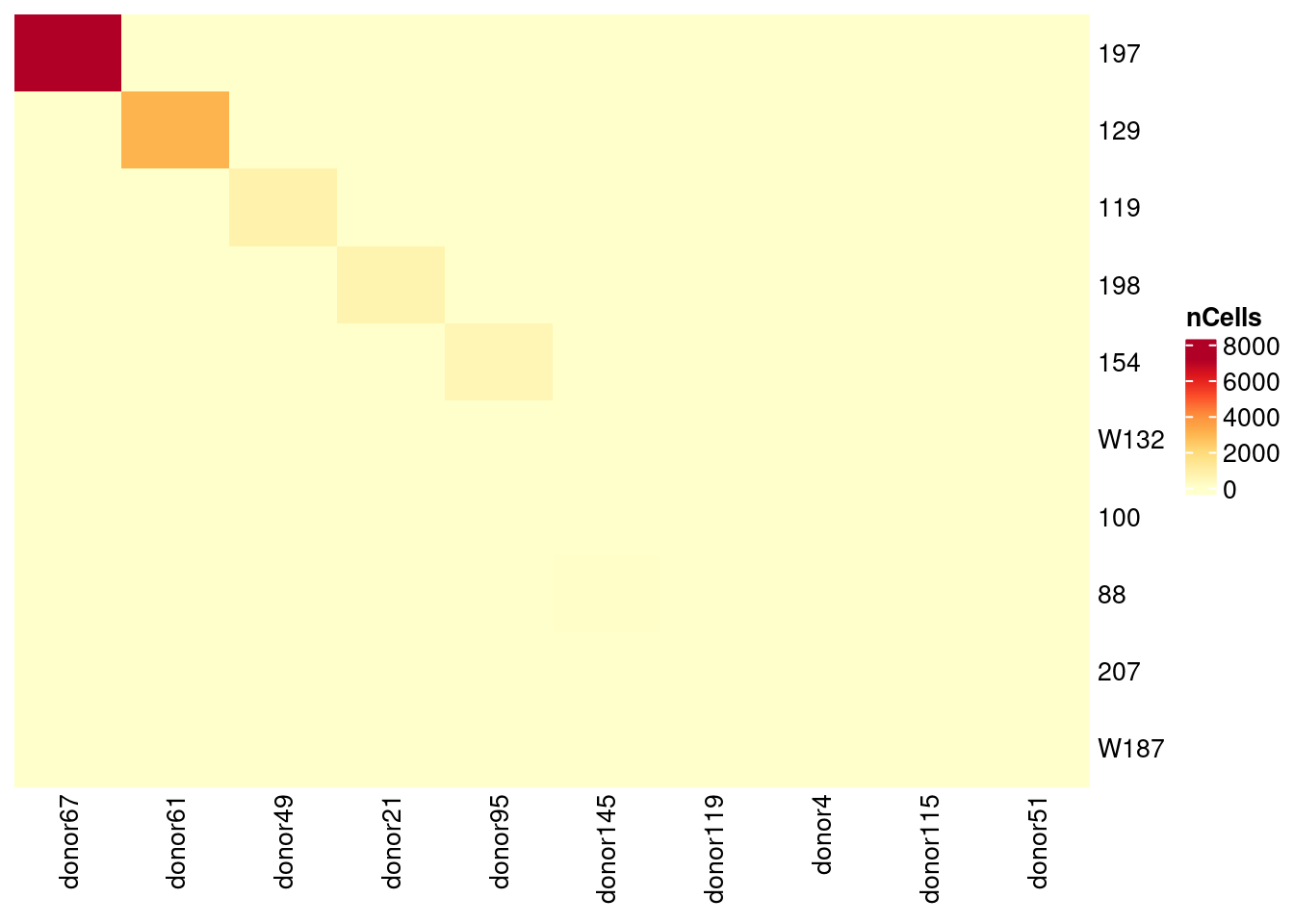
| Version | Author | Date |
|---|---|---|
| c683543 | cazodi | 2022-03-09 |
Vireo vs demuxlet
Vireo will generate most likely additional donors when the requested number of donors is greater than the number of donors in the genotype data (labeled donor136-150), demuxlet does not do this.
donor_ids_assigned <- subset(donor_ids, ! vireo %in% c("unassigned", "doublet"))
vireo_demux <- as.matrix(table(donor_ids_assigned$vireo,
donor_ids_assigned$demuxlet))
Heatmap(log10(1+vireo_demux), name = "log10(nCells+1)",
cluster_rows = FALSE, cluster_columns = FALSE,
column_names_gp = gpar(fontsize = 5), row_names_gp = gpar(fontsize = 5),
col = colorRampPalette(brewer.pal(8, "YlOrRd"))(25))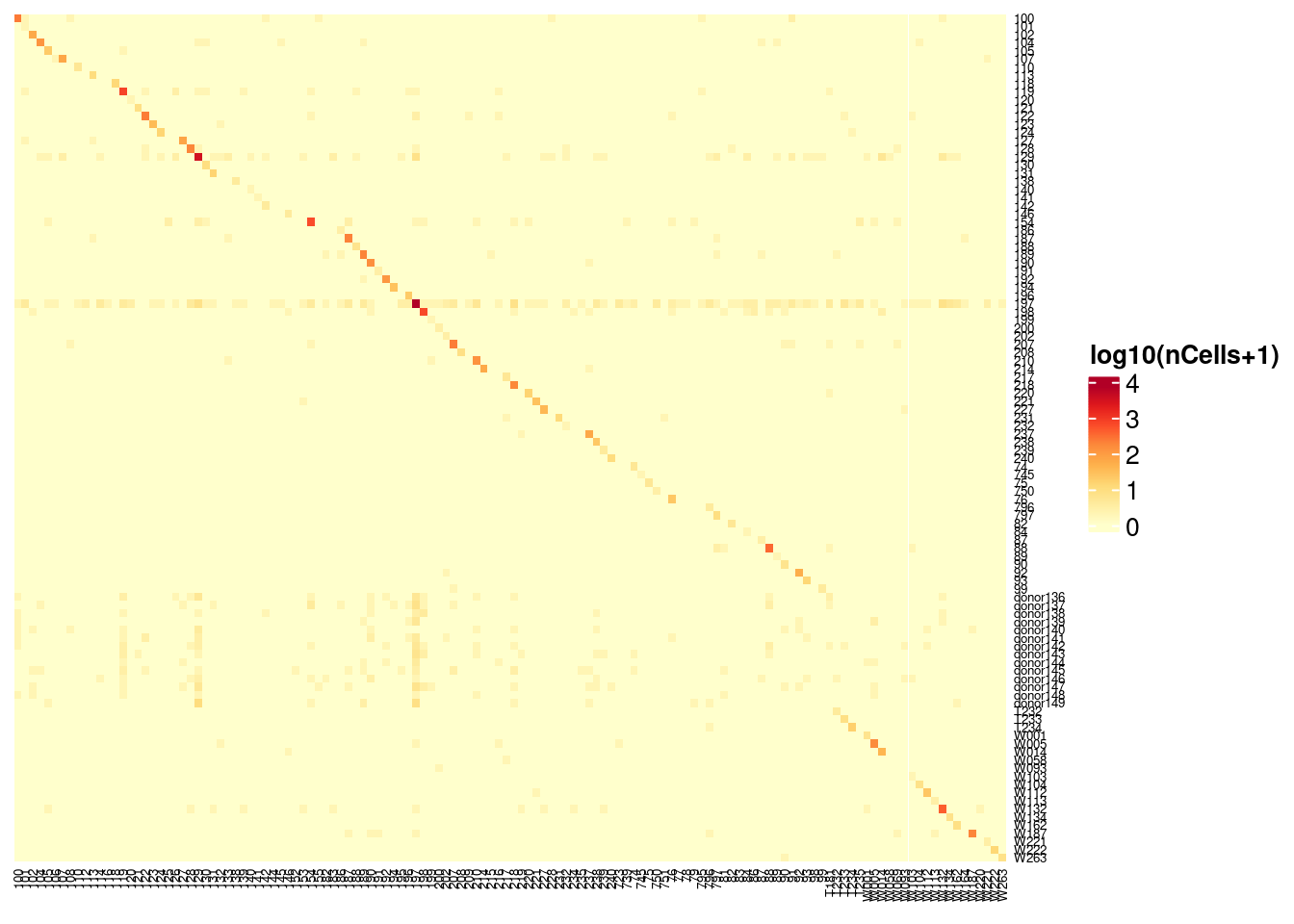
| Version | Author | Date |
|---|---|---|
| c683543 | cazodi | 2022-03-09 |
Vireo GT_barcodes
The GTbarcode function from vireo can be used to generate the minimal set of discriminatory variants. Snapshot of discriminatory variants:
gtbc <- read.table("/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/03_vireo-nCount10/Capture5-GEX/GT_barcodes.tsv",
sep="\t", header=TRUE)
rownames(gtbc) <- gtbc$variants
gtbc$variants <- NULL
gtbc[, c("X101", "X129", "X197", "W221", "donor136")] X101 X129 X197 W221 donor136
15_83035899_A_T 2 1 1 0 0
17_49353734_T_C 2 0 1 0 0
2_117832267_C_T 1 0 1 2 2
12_101296181_A_G 0 1 1 0 2
17_30801092_T_A 1 1 1 0 1
1_85705389_A_G 1 0 0 0 0
7_129051936_A_G 1 2 1 0 0gtbc.cor <- as.matrix(cor(gtbc))
Heatmap(gtbc.cor, name = "cor", column_names_gp = gpar(fontsize = 5),
row_names_gp = gpar(fontsize = 5),
col = colorRampPalette(brewer.pal(8, "RdYlBu"))(25))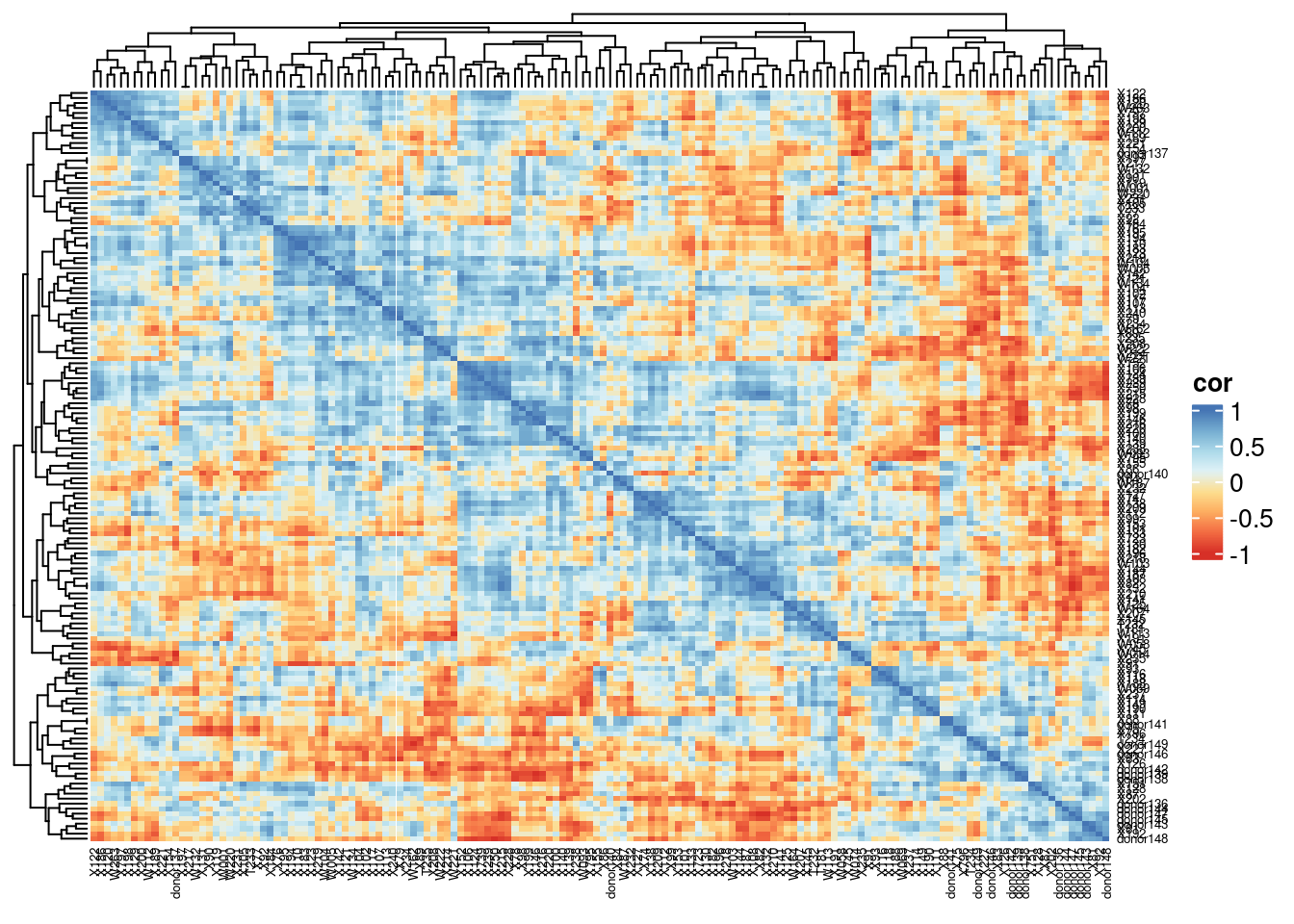
The pairwise correlation between genotypes of the discriminatory variants.
| Version | Author | Date |
|---|---|---|
| c683543 | cazodi | 2022-03-09 |
Vireo by chromosome experiment
wd <- "/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/"
for(chr in 1:22){
tmp <- fread(paste0(wd, "03_vireo/Capture5-GEX_", chr, "/donor_ids.tsv"), sep="\t")
if(chr == 1){
chr_donors <- tmp[, c("cell", "donor_id")]
names(chr_donors) <- c("cell", "chr1")
} else{
tmp <- tmp %>% dplyr::select(cell, donor_id)
names(tmp) <- c("cell", paste0("chr", chr))
chr_donors <- chr_donors %>% left_join(tmp, by="cell")
}
}
# Test chrs 2, 16, 17 together!
tmp <- fread(paste0(wd, "03_vireo-chr2-16-17/donor_ids.tsv"), sep="\t") %>%
dplyr::select(cell, donor_id)
names(tmp) <- c("cell", "chr2-16-17")
chr_donors <- chr_donors %>% left_join(tmp, by="cell")
#
chr_donors2 <- chr_donors %>% mutate(across(where(is.character), ~na_if(., "unassigned")))
chr_donors2 <- chr_donors2[rowSums(is.na(chr_donors2[ , 2:ncol(chr_donors2)])) != 22, ]
chr_donors_stats <- chr_donors %>%
pivot_longer(-cell, names_to = "chromosome", values_to = "donor") %>%
dplyr::filter(donor != "unassigned")
chr_donors_stats2 <- as.matrix(table(chr_donors_stats$chromosome,
chr_donors_stats$donor))
message("Number of assigned cells by chromosome:")Number of assigned cells by chromosome:sort(rowSums(chr_donors_stats2)) chr14 chr20 chr15 chr8 chr10 chr6 chr5
1 1 6 13 15 17 26
chr11 chr9 chr7 chr4 chr12 chr17 chr3
34 35 39 53 59 70 119
chr16 chr19 chr2 chr1 chr2-16-17
129 229 264 351 5330 message("Percent of assigned cells assigned to donor 197:")Percent of assigned cells assigned to donor 197:sort(round(chr_donors_stats2[,"197"] / rowSums(chr_donors_stats2)*100, 1)) chr20 chr5 chr16 chr7 chr2 chr8 chr11
0.0 15.4 17.8 20.5 22.0 23.1 23.5
chr17 chr12 chr2-16-17 chr3 chr1 chr4 chr6
34.3 42.4 44.1 48.7 49.0 50.9 52.9
chr19 chr10 chr9 chr15 chr14
53.7 60.0 74.3 83.3 100.0 chrs_consider <- chr_donors_stats %>% drop_na() %>% group_by(chromosome) %>%
tally() %>% dplyr::filter(n > 50) %>% arrange(desc(-n))
donor_order <- chr_donors_stats %>% drop_na() %>% group_by(donor) %>%
tally() %>% arrange(desc(-n))
chr_donors_stats %>%
drop_na() %>% dplyr::filter(chromosome %in% chrs_consider$chromosome) %>%
mutate(chromosome = gsub("chr", "", chromosome)) %>%
group_by(chromosome, donor) %>%
summarise(cnt = n()) %>%
dplyr::filter(cnt >= 2) %>%
mutate(freq = formattable::percent(cnt / sum(cnt))) %>%
arrange(desc(-freq)) %>%
ggline(x="donor", y="freq", color="chromosome",
order=donor_order$donor) + coord_flip()`summarise()` has grouped output by 'chromosome'. You can override using the
`.groups` argument.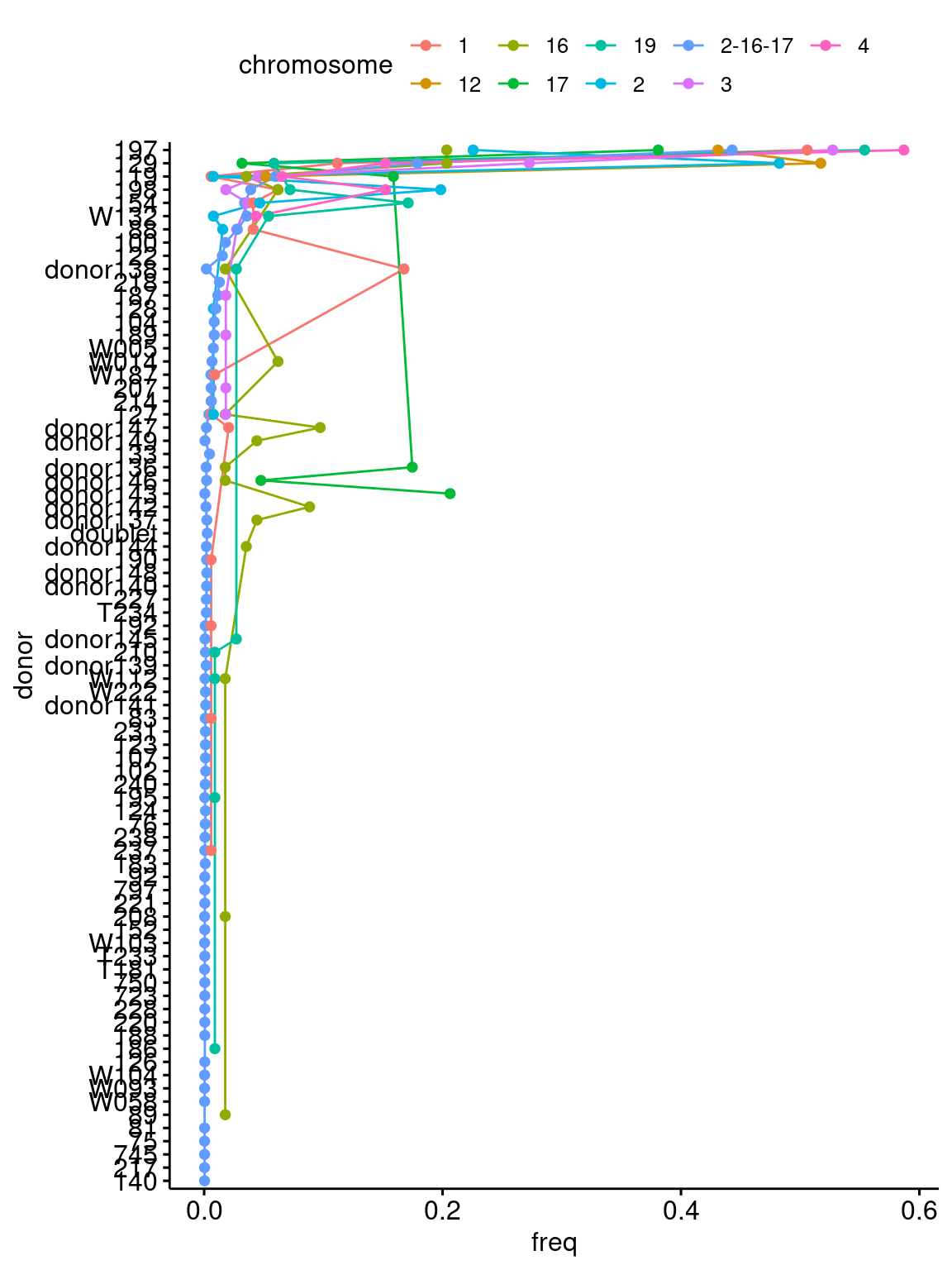
Percent of cells assigned to each donor using SNPs from single chromosomes (color), chromosomes with fewer than 50 donors assigned are removed for simplicity! Donors are sorted by total number of cells assigned across all chromosomes.
| Version | Author | Date |
|---|---|---|
| d01f3b8 | cazodi | 2022-03-30 |
Very consistent that most cells are assigned to donor 197 , donor 129 is a bit less consistent, with some chrs SNPs not assigning may cells there (e.g. chr 17 - which only assigned 70 donors).
SNP filtering
dir <- "/mnt/beegfs/mccarthy/backed_up/general/cazodi/Projects/BAUH_2020_MND-single-cell/output/pilot3.0_iPSC/"
v <- fread(paste0(dir, "03_vireo/Capture5-GEX/donor_ids.tsv"), sep="\t")
v_rmGT7 <- fread(paste0(dir, "03_vireo-rm-vireoGT7/donor_ids.tsv"), sep="\t")
v_max1k <- fread(paste0(dir, "03_vireo-AD_max_1k/donor_ids.tsv"), sep="\t")
v_max500 <- fread(paste0(dir, "03_vireo-AD_max_500/donor_ids.tsv"), sep="\t")
v_min50 <- fread(paste0(dir, "03_vireo-AD_min_50/donor_ids.tsv"), sep="\t")
v_min50max1k <- fread(paste0(dir, "03_vireo-AD_min_50_max_1k/donor_ids.tsv"), sep="\t")
#v_ProtCod <- fread(paste0(dir, "03_vireo-03_vireo-protCod-autos/donor_ids.tsv"), sep="\t")
donor_ids <- v[, c("cell", "donor_id")]
names(donor_ids) <- c("cell", "original")
donor_ids$rmGT7 <- v_rmGT7$donor_id
donor_ids$max1k <- v_max1k$donor_id
donor_ids$max500 <- v_max500$donor_id
donor_ids$min50 <- v_min50$donor_id
donor_ids$min50max1k <- v_min50max1k$donor_id
#donor_ids$protCode <- v_ProtCod$donor_id
head(donor_ids) cell original rmGT7 max1k max500 min50 min50max1k
1: AAACCCAAGCTCGAAG-1 197 197 197 197 197 197
2: AAACCCAAGGAAAGTG-1 129 129 129 129 129 129
3: AAACCCAAGGACTATA-1 154 154 154 154 154 154
4: AAACCCAAGGGTACAC-1 197 197 197 197 197 197
5: AAACCCAAGGTGCTAG-1 100 100 100 100 100 100
6: AAACCCAAGGTTAAAC-1 197 197 197 197 197 unassigned- max1k: removed 235 SNPs, 55,936 remain
- max500: removed 501 SNPs, 55,670 remain
- min50: remove 48,458 SNPs, 7,713 remain
- min50max1k: remove 48,693 SNPs, 7,478 remain
- rmGT7: remove 7 SNPs, 56,164 remain
- protein coding: remove 9,831 SNPs, 46,340 remain
filt_donors_stats <- donor_ids %>%
pivot_longer(-cell, names_to = "test", values_to = "donor") %>%
dplyr::filter(! donor %in% c("doublet", "unassigned"))
filt_donors_stats2 <- as.matrix(table(filt_donors_stats$test,
filt_donors_stats$donor))
message("Number of assigned cells by test:")Number of assigned cells by test:sort(rowSums(filt_donors_stats2))min50max1k min50 max500 max1k rmGT7 original
13246 16748 16910 17152 17509 17563 message("Percent of assigned cells assigned to donor 197:")Percent of assigned cells assigned to donor 197:sort(round(filt_donors_stats2[,"197"] / rowSums(filt_donors_stats2)*100, 1)) min50 original rmGT7 max500 max1k min50max1k
43.8 44.2 44.2 44.7 45.0 45.4 donor_order <- filt_donors_stats %>% drop_na() %>% group_by(donor) %>%
tally() %>% arrange(desc(-n))
filt_donors_stats %>%
drop_na() %>%
group_by(test, donor) %>%
summarise(cnt = n()) %>%
dplyr::filter(cnt >= 10) %>%
mutate(freq = formattable::percent(cnt / sum(cnt))) %>%
arrange(desc(-freq)) %>%
ggline(x="donor", y="freq", color="test",
order=donor_order$donor) + coord_flip()`summarise()` has grouped output by 'test'. You can override using the `.groups`
argument.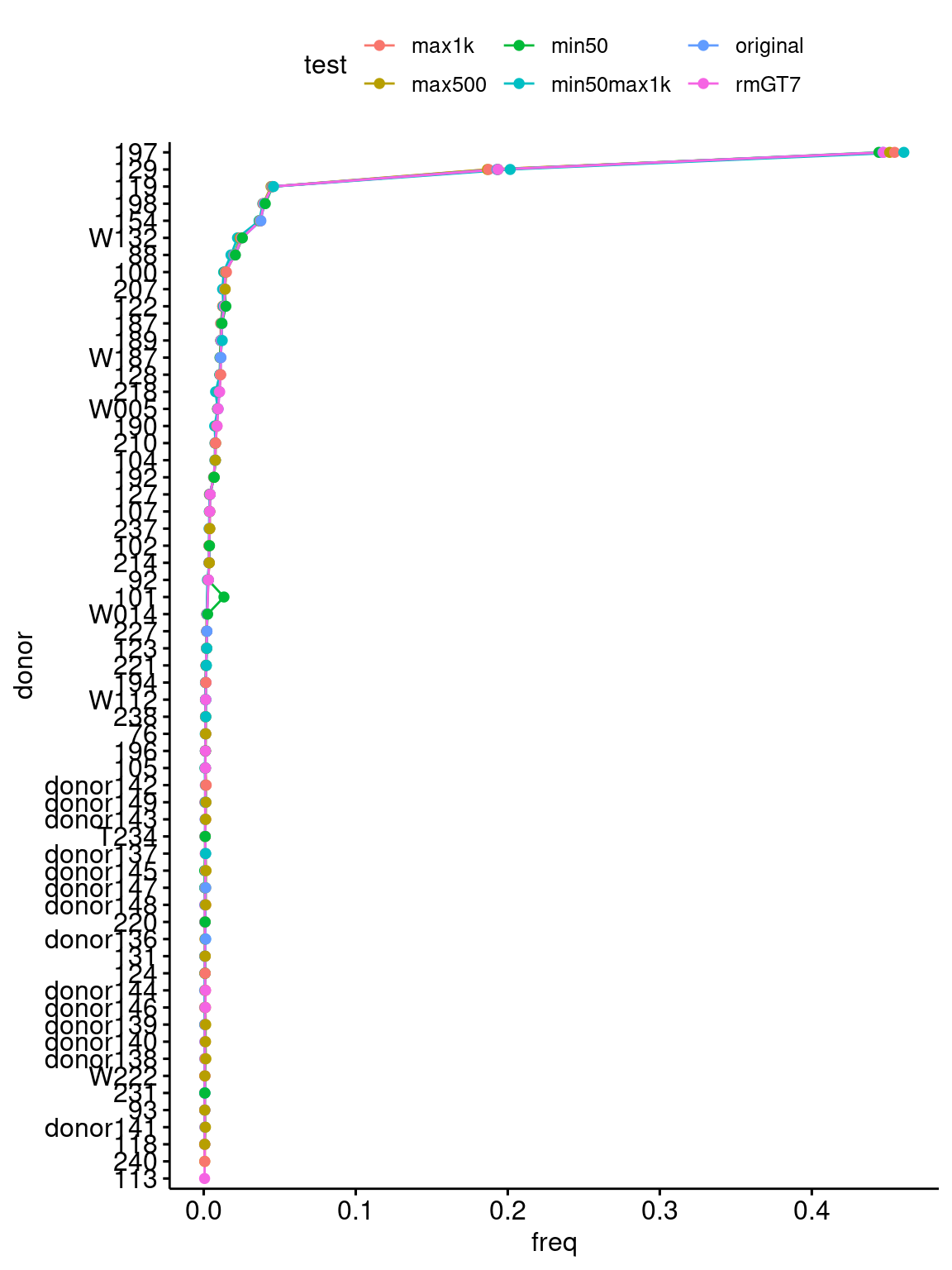
Percent of cells assigned to each donor using different subsets of snps (donors with fewer than 10 cells are removed for simplicity).
| Version | Author | Date |
|---|---|---|
| d01f3b8 | cazodi | 2022-03-30 |
sessionInfo()R version 4.1.1 (2021-08-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Rocky Linux 8.5 (Green Obsidian)
Matrix products: default
BLAS/LAPACK: /usr/lib64/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ggpubr_0.4.0 cowplot_1.1.1 data.table_1.14.2
[4] ComplexHeatmap_2.10.0 RColorBrewer_1.1-2 forcats_0.5.1
[7] stringr_1.4.0 purrr_0.3.4 readr_2.1.1
[10] tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.5
[13] tidyverse_1.3.1 dplyr_1.0.7 corrplot_0.92
loaded via a namespace (and not attached):
[1] matrixStats_0.61.0 fs_1.5.2 lubridate_1.8.0
[4] doParallel_1.0.17 httr_1.4.2 rprojroot_2.0.2
[7] tools_4.1.1 backports_1.4.1 bslib_0.3.1
[10] utf8_1.2.2 R6_2.5.1 DBI_1.1.2
[13] BiocGenerics_0.40.0 colorspace_2.0-3 GetoptLong_1.0.5
[16] withr_2.5.0 tidyselect_1.1.2 compiler_4.1.1
[19] git2r_0.29.0 cli_3.2.0 rvest_1.0.2
[22] xml2_1.3.3 labeling_0.4.2 sass_0.4.0
[25] scales_1.1.1 digest_0.6.29 rmarkdown_2.11
[28] pkgconfig_2.0.3 htmltools_0.5.2 dbplyr_2.1.1
[31] fastmap_1.1.0 highr_0.9 htmlwidgets_1.5.4
[34] rlang_1.0.2 GlobalOptions_0.1.2 readxl_1.3.1
[37] rstudioapi_0.13 farver_2.1.0 shape_1.4.6
[40] jquerylib_0.1.4 generics_0.1.2 jsonlite_1.8.0
[43] car_3.0-12 magrittr_2.0.2 Rcpp_1.0.8
[46] munsell_0.5.0 S4Vectors_0.32.3 fansi_1.0.0
[49] abind_1.4-5 lifecycle_1.0.1 stringi_1.7.6
[52] whisker_0.4 yaml_2.3.5 carData_3.0-5
[55] parallel_4.1.1 promises_1.2.0.1 crayon_1.5.0
[58] haven_2.4.3 circlize_0.4.14 hms_1.1.1
[61] magick_2.7.3 knitr_1.37 pillar_1.6.4
[64] rjson_0.2.21 ggsignif_0.6.3 codetools_0.2-18
[67] stats4_4.1.1 reprex_2.0.1 glue_1.6.0
[70] evaluate_0.15 modelr_0.1.8 png_0.1-7
[73] vctrs_0.3.8 tzdb_0.2.0 httpuv_1.6.5
[76] foreach_1.5.2 cellranger_1.1.0 gtable_0.3.0
[79] clue_0.3-60 assertthat_0.2.1 formattable_0.2.1
[82] xfun_0.30 broom_0.7.10 rstatix_0.7.0
[85] later_1.3.0 iterators_1.0.14 IRanges_2.28.0
[88] cluster_2.1.2 workflowr_1.6.2 ellipsis_0.3.2