Misc: Co-regulation
Christina B. Azodi
2021-09-23
Last updated: 2021-09-23
Checks: 6 1
Knit directory: KEJP_2020_splatPop/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210215) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f2bf14a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/10x_lung/
Ignored: data/ALL.chr2.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.EURO.0.99.MAF05.filtered.vcf
Ignored: data/D30.h5
Ignored: data/Diabetes/
Ignored: data/IBD/
Ignored: data/agg_10X-fibroblasts-control.rds
Ignored: data/agg_IBD-ss2.rds
Ignored: data/agg_Neuro-10x_DA.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6.rds
Ignored: data/agg_diabetes-ss2.rds
Ignored: data/agg_iPSC-ss2_D0.rds
Ignored: data/covid/
Ignored: data/cuomo_NeuroSeq_10x_all_sce.rds
Ignored: data/iPSC_eqtl-mapping/
Ignored: data/lung_fibrosis/
Ignored: data/pseudoB_IBD-ss2.rds
Ignored: data/pseudoB_Neuro-10x_DA.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6.rds
Ignored: data/pseudoB_diabetes-ss2.rds
Ignored: data/pseudoB_iPSC-ss2_D0.rds
Ignored: data/sce_10X-fibroblasts-947170.rds
Ignored: data/sce_10X-fibroblasts-allGenes.rds
Ignored: data/sce_10X-fibroblasts.rds
Ignored: data/sce_IBD-ss2.rds
Ignored: data/sce_IBD-ss2_HC2.rds
Ignored: data/sce_Neuro-10x.rds
Ignored: data/sce_Neuro-10x_2CT.rds
Ignored: data/sce_Neuro-10x_DA-wihj4.rds
Ignored: data/sce_Neuro-10x_DA_allGenes.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6.rds
Ignored: data/sce_diabetes-ss2.rds
Ignored: data/sce_diabetes-ss2_T2D-5.rds
Ignored: data/sce_iPSC-ss2_D0-allGenes.rds
Ignored: data/sce_iPSC-ss2_D0-joxm39.rds
Ignored: data/sce_iPSC-ss2_D0.rds
Ignored: output/00_Figures/
Ignored: output/01_sims/
Ignored: output/10x_eQTL-mapping/
Ignored: output/demo_eQTL/
Ignored: output/iPSC_eQTL/
Ignored: references/1000GP_Phase3_sample_info.txt
Ignored: references/Homo_sapiens.GRCh38.99.chromosome.22.gff3
Ignored: references/chr2.filt.2-temporary.bed
Ignored: references/chr2.filt.2-temporary.bim
Ignored: references/chr2.filt.2-temporary.fam
Ignored: references/chr2.filt.2.log
Ignored: references/chr2.filt.log
Ignored: references/chr2.filt.map
Ignored: references/chr2.filt.nosex
Ignored: references/chr2.filt.ped
Ignored: references/chr2.filt.prune.in
Ignored: references/chr2.filt.prune.out
Ignored: references/chr2.filtered.log
Ignored: references/chr2.filtered.nosex
Ignored: references/chr2.filtered.vcf
Ignored: references/chr2.filtered2.log
Ignored: references/chr2.filtered2.nosex
Ignored: references/chr2.filtered2.vcf
Ignored: references/chr2.filtered3.log
Ignored: references/chr2.filtered3.nosex
Ignored: references/chr2.filtered3.vcf
Ignored: references/chr2.genes.gff3
Ignored: references/chr2.vcf.gz
Ignored: references/chr22.filt.log
Ignored: references/chr22.filt.map
Ignored: references/chr22.filt.nosex
Ignored: references/chr22.filt.ped
Ignored: references/chr22.filt.prune.in
Ignored: references/chr22.filt.prune.out
Ignored: references/chr22.filtered.log
Ignored: references/chr22.filtered.nosex
Ignored: references/chr22.filtered.vcf
Ignored: references/chr22.filtered.vcf.bed
Ignored: references/chr22.filtered.vcf.bim
Ignored: references/chr22.filtered.vcf.fam
Ignored: references/chr22.filtered.vcf.log
Ignored: references/chr22.filtered.vcf.nosex
Ignored: references/chr22.filtered.vcf.rel
Ignored: references/chr22.filtered.vcf.rel.id
Ignored: references/chr22.filtered.vcf.rel_mod
Ignored: references/chr22.genes.gff3
Ignored: references/chr22.genes.gff3_annotation
Ignored: references/chr22.genes.gff3_chunks
Ignored: references/chr22.vcf.gz
Ignored: references/eqtl_summary_stats.tar.gz
Ignored: references/eqtl_summary_stats_renamed/
Ignored: references/keep_samples.txt
Ignored: references/remove_snps.txt
Ignored: references/test.genome
Ignored: references/test.log
Ignored: references/test.nosex
Untracked files:
Untracked: .DS_Store
Untracked: .cache/
Untracked: .config/
Untracked: .snakemake/
Untracked: KEJP_2020_splatPop.Rproj
Untracked: code/splatter_1.17.2.tar.gz
Untracked: docs/.DS_Store
Unstaged changes:
Modified: .gitignore
Modified: .gitlab-ci.yml
Modified: CITATION
Modified: Dockerfile
Modified: LICENSE
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/10x-Neuro_emp-simulations.Rmd
Modified: analysis/10x-Neuro_estimate-params.Rmd
Modified: analysis/KEJP_iPSC-ss2.Rmd
Modified: analysis/_site.yml
Modified: analysis/about.Rmd
Modified: analysis/index.Rmd
Modified: analysis/license.Rmd
Modified: analysis/misc_analysis-coregulation.Rmd
Modified: analysis/ss2-iPSC_estimate-params.Rmd
Modified: analysis/ss2-iPSC_simulations.Rmd
Modified: cluster.json
Modified: code/1_process-empirical-data.R
Modified: code/2_estimate-splatPopParams.R
Modified: code/4_simulate_DEG.R
Modified: code/README.md
Modified: code/plot_functions.R
Modified: code/plot_functions2.R
Modified: data/README.md
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.css
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.js
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cerulean.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cosmo.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/darkly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/flatly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Lato.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycle.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycleBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSans.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBoldItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLightItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Raleway.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RalewayBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Roboto.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoMedium.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansPro.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Ubuntu.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/journal.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/lumen.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/paper.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/readable.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/sandstone.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/simplex.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/spacelab.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/united.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/yeti.min.css
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.eot
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.svg
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.ttf
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff2
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.js
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.min.js
Modified: docs/site_libs/bootstrap-3.3.5/js/npm.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/html5shiv.min.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/respond.min.js
Modified: docs/site_libs/font-awesome-5.1.0/css/all.css
Modified: docs/site_libs/font-awesome-5.1.0/css/v4-shims.css
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff2
Modified: docs/site_libs/header-attrs-2.5/header-attrs.js
Modified: docs/site_libs/jquery-1.11.3/jquery.min.js
Modified: docs/site_libs/jqueryui-1.11.4/README
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_444444_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_555555_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777620_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777777_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_cc0000_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_ffffff_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/index.html
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.min.css
Modified: docs/site_libs/navigation-1.1/codefolding.js
Modified: docs/site_libs/navigation-1.1/sourceembed.js
Modified: docs/site_libs/navigation-1.1/tabsets.js
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.css
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.js
Modified: environment.yml
Modified: envs/limix_env.yaml
Modified: envs/myenv.yaml
Modified: org/README.md
Modified: org/project_management.org
Modified: output/README.md
Modified: references/README.md
Modified: resources/README.md
Modified: resources/keep_samples.txt
Modified: workflow/config_bp_ss2.yaml
Modified: workflow/sims_limix_v1.15.1.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/misc_analysis-coregulation.Rmd) and HTML (public/misc_analysis-coregulation.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9effdc2 | cazodi | 2021-08-31 | added coregulation example |
| html | 9effdc2 | cazodi | 2021-08-31 | added coregulation example |
suppressPackageStartupMessages({
library(splatter)
library(VariantAnnotation)
library(tidyverse)
library(ggpubr)
library(cowplot)
})
source("code/plot_functions.R")
source("code/misc_functions.R")
rerun <- FALSE
save <- TRUE
date <- Sys.Date()
date.use <- "2021-09-23"
set.seed(42)Load 10x-Neuro data
# Chromosome 22 data
gff <- read.table("references/chr22.genes.gff3", sep="\t", header=FALSE, quote="")
vcf <- readVcf("references/chr22.filtered.vcf", "hg38")
sampleNames <- colnames(geno(vcf)$GT)
# Smartseq2 iPSC data and splatPopParams
sce <- readRDS("data/sce_Neuro-10x_2CT.rds")
sce$Batch <- "Batch1"
params <- readRDS("output/01_sims/splatPop-params_Neuro-10x.rds")Gene co-regulation
splatPop randomly selects an eSNP within the desired window for each eGene. By chance some eGenes may be assigned the same eSNP, but if many SNPs are provided this is rare. However, the eqtl.coreg parameter can be used to randomly select a percent of eGenes, sort them by chromosome and location, and assign them the same eSNP. This function maintains the randomly sampled effect sizes and designation as a global, group specific, and/or condition specific eQTL, however it does ensure that all eQTL with the same eSNP are given the same effect sign. Here is an example of a splatPop key with coregulation effects, sorted by eSNP so you can see some co-regulated genes:
vcf100 <- vcf[, sample(colnames(vcf), 100)]
if(rerun) {
params.coreg <- setParams(params, eqtl.coreg = 0.2, eqtl.n=1)
sim.coreg <- splatPopSimulateMeans(vcf = vcf100, gff = gff,
params = params.coreg)
if(save){
save.name <- paste0(date, "_coreg-10x")
saveRDS(sim.coreg, paste0("output/01_sims/", save.name, ".rds"))
}
} else{
sim.coreg <- readRDS(paste0("output/01_sims/", date.use, "_coreg-10x.rds"))
}
key <- sim.coreg$key
head(arrange(key, desc(eSNP.ID)), n = 10) geneID chromosome geneStart geneEnd geneMiddle
1 ENSG00000063515 22 19148576 19150283 19149429
2 ENSG00000278881 22 47115838 47117217 47116527
3 ENSG00000128311 22 37010859 37020183 37015521
4 ENSG00000073146 22 50089879 50161690 50125784
5 ENSG00000056487 22 44881162 45010005 44945583
6 ENSG00000159873 22 28772674 28789301 28780987
7 ENSG00000211685 22 22922594 22923034 22922814
8 ENSG00000242247 22 42796502 42858106 42827304
9 ENSG00000211648 22 22357739 22358260 22357999
10 ENSG00000179750 22 38982347 38992804 38987575
meanSampled.noOutliers OutlierFactor meanSampled cvSampled eQTL.group
1 1.416049564 1.0000 1.416049564 0.12870805 global
2 0.193234111 133.7316 24.620750940 0.09012078 global
3 1.968574357 1.0000 1.968574357 0.18191912 global
4 1.420318754 1.0000 1.420318754 0.11297984 global
5 0.009623997 1.0000 0.009623997 0.76205240 global
6 0.932489055 1.0000 0.932489055 0.31186934 global
7 0.036344559 1.0000 0.036344559 0.38613589 global
8 0.327289877 1.0000 0.327289877 0.46399448 global
9 2.705201728 1.0000 2.705201728 0.19001227 global
10 0.027653153 1.0000 0.027653153 0.35767593 global
eQTL.condition eSNP.ID eSNP.chromosome eSNP.loc eSNP.MAF eQTL.EffectSize
1 global rs9808864 22 19646034 0.240 -0.28989508
2 global rs9680608 22 46856102 0.470 0.35121750
3 global rs963621 22 37555766 0.470 -0.28325193
4 global rs9627711 22 49769873 0.300 -0.04800535
5 global rs9626485 22 44940564 0.085 0.17811913
6 global rs9625262 22 27958126 0.265 -0.19831520
7 global rs9624040 22 23509902 0.420 -0.48831633
8 global rs9623589 22 42817652 0.255 0.25915278
9 global rs9622981 22 22504493 0.315 0.41421010
10 global rs9622770 22 38674460 0.450 -0.47521664
ConditionDE.Condition1
1 1
2 1
3 1
4 1
5 1
6 1
7 1
8 1
9 1
10 1We can compare the expression correlation between randomly paired genes in a splatPop simulation (grey) with the correlation between co-regulated genes (blue), with the average correlation for each type designated by the large point.
sim.cor <- cor(t(sim.coreg$means))
coregSNPs <- names(which(table(sim.coreg$key$eSNP.ID) == 2))
coregCorList <- c()
coregESMax <- c()
coregESMin <- c()
coregMAFList <- c()
for(snp in coregSNPs){
coregGenes <- sim.coreg$key$geneID[which(sim.coreg$key$eSNP.ID == snp)]
coregCorList <- c(coregCorList, sim.cor[coregGenes[1], coregGenes[2]])
coregESMax <- c(coregESMax, max(key[key$geneID %in% coregGenes,
"eQTL.EffectSize"]))
coregESMin <- c(coregESMin, min(key[key$geneID %in% coregGenes,
"eQTL.EffectSize"]))
coregMAFList <- c(coregMAFList, key[key$geneID == coregGenes[1], "eSNP.MAF"])
}
randCorList <- c()
reps <- 1000
for (n in 1:reps){
genes <- sample(key$geneID, 2)
randCorList <- c(randCorList, sim.cor[genes[1], genes[2]])
}
cor.df <- as.data.frame(list(type=c(rep("coregulated pairs", length(coregCorList)),
rep("random pairs", length(randCorList))),
correlation=c(coregCorList, randCorList),
effect_size_max = c(abs(coregESMax), rep(NA, reps)),
effect_size_min = c(abs(coregESMin), rep(NA, reps)),
maf = c(coregMAFList, rep(NA, reps)),
id = "x"))
ggplot(cor.df, aes(x = correlation, y = id, color = type, alpha = type)) +
geom_jitter(size = 1.5, width = 0.1) +
scale_alpha_discrete(range = c(0.5, 0.1)) +
stat_summary(fun = mean, geom = "pointrange", size = 1, alpha=1) + theme_minimal() +
theme(axis.title.y=element_blank(), axis.text.y=element_blank(),
legend.position="top") + xlim(c(-1, 1)) +
scale_color_manual(values=c("#56B4E9", "#999999"))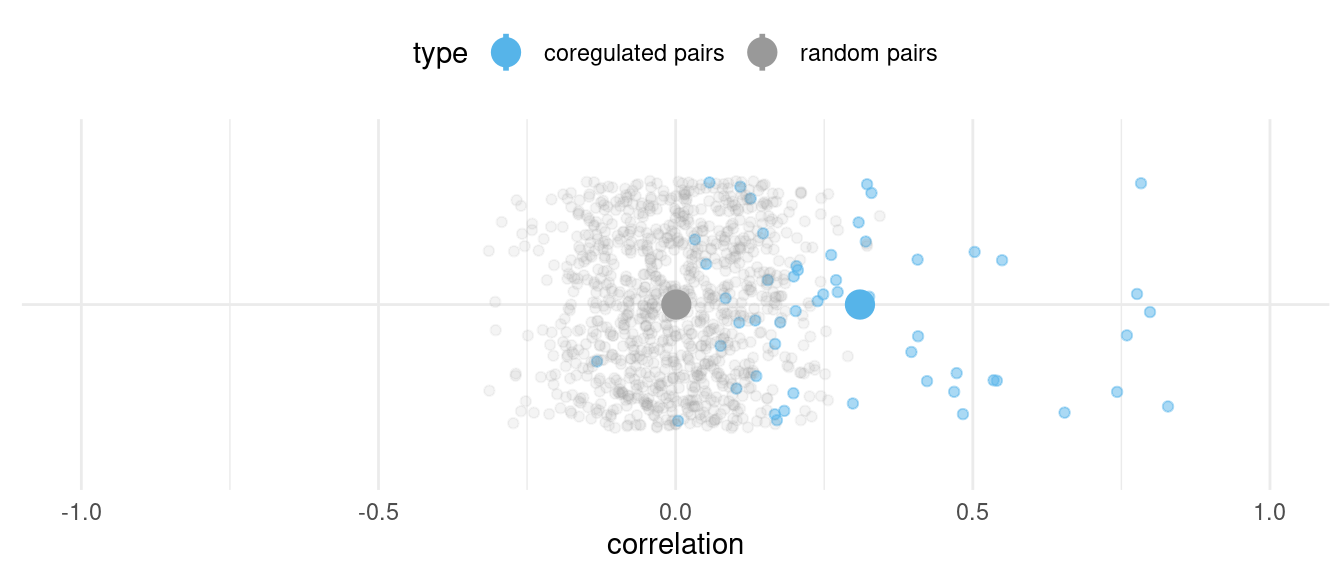
Mean aggregated gene expression correlation between randomly paired (grey) and co-regulated (blue) genes across 100 individuals, with the average correlation for each type designated by the large point.
| Version | Author | Date |
|---|---|---|
| 9effdc2 | cazodi | 2021-08-31 |
if(save){
save.name <- paste0(date, "_coreg")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 5, height = 2)
}p1 <- cor.df %>% filter(type == "coregulated pairs") %>%
ggscatter(x="effect_size_min", y="effect_size_max", color="correlation",
xlim=c(0.1,0.7), ylim=c(0.1,0.7)) +
geom_abline(intercept = 0, slope = 1) +
scale_colour_gradient(low="yellow", high= "red")
p2 <- cor.df %>% filter(type == "coregulated pairs") %>%
ggscatter(x="maf", y="correlation", color="correlation") +
scale_colour_gradient(low="yellow", high= "red")
ggarrange(p1, p2, common.legend = TRUE)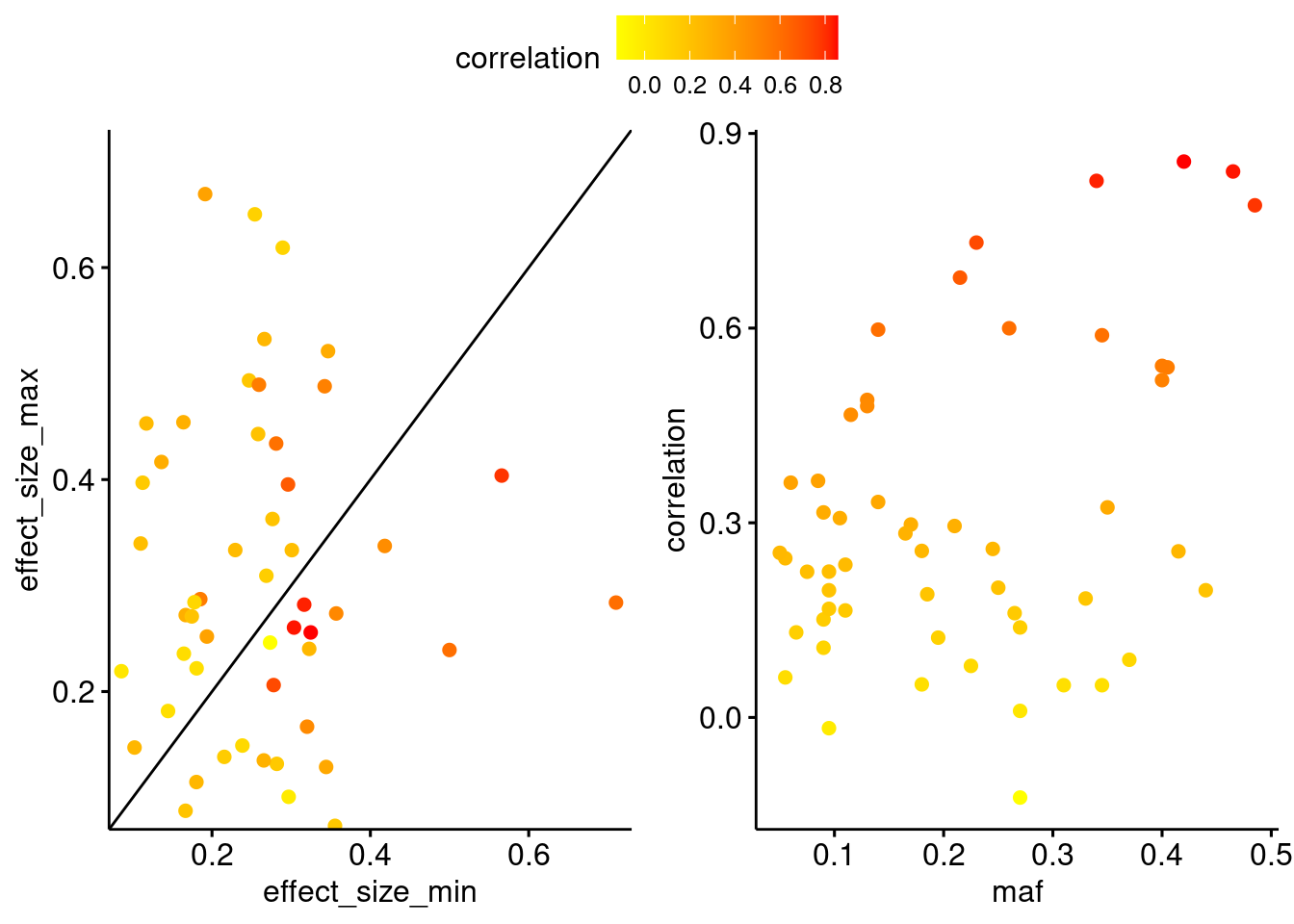
Characteristics of coregulated genes by the degree of gene expression correlation. Left plot shows the simulated eQTL effect size for each pair (x-axis shows the minimum and y-axis shows the maximum), with genes with higher expression correlation in red. Genes pairs along the diagonal represent those where the same eSNP has a similar effect size on both genes. Right plot shows the relationship between the minor allele frequency of the eSNP and the gene expression correlation.
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.0.4 (2021-02-15)
os Red Hat Enterprise Linux 8.4 (Ootpa)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-09-23
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.0.2)
AnnotationDbi 1.52.0 2020-10-27 [1] Bioconductor
askpass 1.1 2019-01-13 [1] CRAN (R 4.0.2)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.2)
backports 1.2.1 2020-12-09 [1] CRAN (R 4.0.4)
Biobase * 2.50.0 2020-10-27 [1] Bioconductor
BiocFileCache 1.14.0 2020-10-27 [1] Bioconductor
BiocGenerics * 0.36.1 2021-04-16 [1] Bioconductor
BiocParallel 1.24.1 2020-11-06 [1] Bioconductor
biomaRt 2.46.3 2021-02-09 [1] Bioconductor
Biostrings * 2.58.0 2020-10-27 [1] Bioconductor
bit 4.0.4 2020-08-04 [1] CRAN (R 4.0.2)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.0.2)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.0.4)
blob 1.2.2 2021-07-23 [1] CRAN (R 4.0.4)
broom 0.7.9 2021-07-27 [1] CRAN (R 4.0.4)
BSgenome 1.58.0 2020-10-27 [1] Bioconductor
bslib 0.2.5.1 2021-05-18 [1] CRAN (R 4.0.4)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.4)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.0.4)
car 3.0-11 2021-06-27 [1] CRAN (R 4.0.4)
carData 3.0-4 2020-05-22 [1] CRAN (R 4.0.2)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.0.2)
checkmate 2.0.0 2020-02-06 [1] CRAN (R 4.0.2)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.0.4)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.0.4)
cowplot * 1.1.1 2020-12-30 [1] CRAN (R 4.0.4)
crayon 1.4.1 2021-02-08 [1] CRAN (R 4.0.4)
curl 4.3.2 2021-06-23 [1] CRAN (R 4.0.4)
data.table 1.14.0 2021-02-21 [1] CRAN (R 4.0.4)
DBI 1.1.1 2021-01-15 [1] CRAN (R 4.0.4)
dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.0.4)
DelayedArray 0.16.3 2021-03-24 [1] Bioconductor
desc 1.3.0 2021-03-05 [1] CRAN (R 4.0.4)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.0.4)
digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.2)
dplyr * 1.0.7 2021-06-18 [1] CRAN (R 4.0.4)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.0.4)
evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.2)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.0.4)
farver 2.1.0 2021-02-28 [1] CRAN (R 4.0.4)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.0.3)
forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.0.4)
foreign 0.8-81 2020-12-22 [2] CRAN (R 4.0.4)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
GenomeInfoDb * 1.26.7 2021-04-08 [1] Bioconductor
GenomeInfoDbData 1.2.4 2020-11-10 [1] Bioconductor
GenomicAlignments 1.26.0 2020-10-27 [1] Bioconductor
GenomicFeatures 1.42.3 2021-04-01 [1] Bioconductor
GenomicRanges * 1.42.0 2020-10-27 [1] Bioconductor
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.0.4)
ggpubr * 0.4.0 2020-06-27 [1] CRAN (R 4.0.3)
ggsignif 0.6.2 2021-06-14 [1] CRAN (R 4.0.4)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.0.4)
glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.0.2)
gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.2)
haven 2.4.3 2021-08-04 [1] CRAN (R 4.0.4)
highr 0.9 2021-04-16 [1] CRAN (R 4.0.4)
hms 1.1.0 2021-05-17 [1] CRAN (R 4.0.4)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.0.4)
httpuv 1.6.2 2021-08-18 [1] CRAN (R 4.0.4)
httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
IRanges * 2.24.1 2020-12-12 [1] Bioconductor
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.4)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.4)
knitr 1.33 2021-04-24 [1] CRAN (R 4.0.4)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.0.2)
later 1.3.0 2021-08-18 [1] CRAN (R 4.0.4)
lattice 0.20-41 2020-04-02 [2] CRAN (R 4.0.4)
lifecycle 1.0.0 2021-02-15 [1] CRAN (R 4.0.4)
locfit 1.5-9.4 2020-03-25 [1] CRAN (R 4.0.2)
lubridate 1.7.10 2021-02-26 [1] CRAN (R 4.0.4)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.3)
Matrix 1.3-4 2021-06-01 [1] CRAN (R 4.0.4)
MatrixGenerics * 1.2.1 2021-01-30 [1] Bioconductor
matrixStats * 0.60.0 2021-07-26 [1] CRAN (R 4.0.4)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.4)
modelr 0.1.8 2020-05-19 [1] CRAN (R 4.0.2)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.2)
openssl 1.4.4 2021-04-30 [1] CRAN (R 4.0.4)
openxlsx 4.2.4 2021-06-16 [1] CRAN (R 4.0.4)
pillar 1.6.2 2021-07-29 [1] CRAN (R 4.0.4)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.4)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
pkgload 1.2.1 2021-04-06 [1] CRAN (R 4.0.4)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.2)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.0.4)
progress 1.2.2 2019-05-16 [1] CRAN (R 4.0.2)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.0.4)
ps 1.6.0 2021-02-28 [1] CRAN (R 4.0.4)
purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.0.4)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.0.4)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.0.4)
RCurl 1.98-1.4 2021-08-17 [1] CRAN (R 4.0.4)
readr * 2.0.1 2021-08-10 [1] CRAN (R 4.0.4)
readxl 1.3.1 2019-03-13 [1] CRAN (R 4.0.2)
remotes 2.4.0 2021-06-02 [1] CRAN (R 4.0.4)
reprex 2.0.1 2021-08-05 [1] CRAN (R 4.0.4)
rio 0.5.27 2021-06-21 [1] CRAN (R 4.0.4)
rlang 0.4.11 2021-04-30 [1] CRAN (R 4.0.4)
rmarkdown 2.10 2021-08-06 [1] CRAN (R 4.0.4)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.3)
Rsamtools * 2.6.0 2020-10-27 [1] Bioconductor
RSQLite 2.2.8 2021-08-21 [1] CRAN (R 4.0.4)
rstatix 0.7.0 2021-02-13 [1] CRAN (R 4.0.4)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.3)
rtracklayer 1.50.0 2020-10-27 [1] Bioconductor
rvest 1.0.1 2021-07-26 [1] CRAN (R 4.0.4)
S4Vectors * 0.28.1 2020-12-09 [1] Bioconductor
sass 0.4.0 2021-05-12 [1] CRAN (R 4.0.4)
scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.2)
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
SingleCellExperiment * 1.12.0 2020-10-27 [1] Bioconductor
splatter * 1.17.2 2021-09-20 [1] Bioconductor
stringi 1.7.3 2021-07-16 [1] CRAN (R 4.0.4)
stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
SummarizedExperiment * 1.20.0 2020-10-27 [1] Bioconductor
testthat 3.0.4 2021-07-01 [1] CRAN (R 4.0.4)
tibble * 3.1.3 2021-07-23 [1] CRAN (R 4.0.4)
tidyr * 1.1.3 2021-03-03 [1] CRAN (R 4.0.4)
tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.0.4)
tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 4.0.4)
tzdb 0.1.2 2021-07-20 [1] CRAN (R 4.0.4)
usethis 2.0.1 2021-02-10 [1] CRAN (R 4.0.4)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.4)
VariantAnnotation * 1.36.0 2020-10-27 [1] Bioconductor
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.0.4)
whisker 0.4 2019-08-28 [1] CRAN (R 4.0.2)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.0.4)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.0.2)
xfun 0.25 2021-08-06 [1] CRAN (R 4.0.4)
XML 3.99-0.7 2021-08-17 [1] CRAN (R 4.0.4)
xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.2)
XVector * 0.30.0 2020-10-27 [1] Bioconductor
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
zip 2.2.0 2021-05-31 [1] CRAN (R 4.0.4)
zlibbioc 1.36.0 2020-10-27 [1] Bioconductor
[1] /mnt/mcfiles/cazodi/R/x86_64-pc-linux-gnu-library/4.0
[2] /opt/R/4.0.4/lib/R/library