SmartSeq2 iPSC: Simulations
Christina B. Azodi
2021-11-15
Last updated: 2021-11-15
Checks: 6 1
Knit directory: KEJP_2020_splatPop/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210215) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 116e4a3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/10x_lung/
Ignored: data/ALL.chr2.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.EURO.0.99.MAF05.filtered.vcf
Ignored: data/D30.h5
Ignored: data/Diabetes/
Ignored: data/IBD/
Ignored: data/agg_10X-fibroblasts-control.rds
Ignored: data/agg_IBD-ss2.rds
Ignored: data/agg_Neuro-10x_DA.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6.rds
Ignored: data/agg_diabetes-ss2.rds
Ignored: data/agg_iPSC-ss2_D0.rds
Ignored: data/covid/
Ignored: data/cuomo_NeuroSeq_10x_all_sce.rds
Ignored: data/iPSC_eqtl-mapping/
Ignored: data/lung_fibrosis/
Ignored: data/pseudoB_IBD-ss2.rds
Ignored: data/pseudoB_Neuro-10x_DA.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6.rds
Ignored: data/pseudoB_diabetes-ss2.rds
Ignored: data/pseudoB_iPSC-ss2_D0.rds
Ignored: data/sce_10X-fibroblasts-947170.rds
Ignored: data/sce_10X-fibroblasts-allGenes.rds
Ignored: data/sce_10X-fibroblasts.rds
Ignored: data/sce_IBD-ss2.rds
Ignored: data/sce_IBD-ss2_HC2.rds
Ignored: data/sce_Neuro-10x.rds
Ignored: data/sce_Neuro-10x_2CT.rds
Ignored: data/sce_Neuro-10x_DA-wihj4.rds
Ignored: data/sce_Neuro-10x_DA_allGenes.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6.rds
Ignored: data/sce_diabetes-ss2.rds
Ignored: data/sce_diabetes-ss2_T2D-5.rds
Ignored: data/sce_iPSC-ss2_D0-allGenes.rds
Ignored: data/sce_iPSC-ss2_D0-joxm39.rds
Ignored: data/sce_iPSC-ss2_D0.rds
Ignored: output/00_Figures/
Ignored: output/01_sims/
Ignored: output/10x_eQTL-mapping/
Ignored: output/demo_eQTL/
Ignored: output/iPSC_eQTL/
Ignored: references/1000GP_Phase3_sample_info.txt
Ignored: references/Homo_sapiens.GRCh38.99.chromosome.22.gff3
Ignored: references/chr2.filt.2-temporary.bed
Ignored: references/chr2.filt.2-temporary.bim
Ignored: references/chr2.filt.2-temporary.fam
Ignored: references/chr2.filt.2.log
Ignored: references/chr2.filt.log
Ignored: references/chr2.filt.map
Ignored: references/chr2.filt.nosex
Ignored: references/chr2.filt.ped
Ignored: references/chr2.filt.prune.in
Ignored: references/chr2.filt.prune.out
Ignored: references/chr2.filtered.log
Ignored: references/chr2.filtered.nosex
Ignored: references/chr2.filtered.vcf
Ignored: references/chr2.filtered2.log
Ignored: references/chr2.filtered2.nosex
Ignored: references/chr2.filtered2.vcf
Ignored: references/chr2.filtered3.log
Ignored: references/chr2.filtered3.nosex
Ignored: references/chr2.filtered3.vcf
Ignored: references/chr2.genes.gff3
Ignored: references/chr2.vcf.gz
Ignored: references/chr22.filt.log
Ignored: references/chr22.filt.map
Ignored: references/chr22.filt.nosex
Ignored: references/chr22.filt.ped
Ignored: references/chr22.filt.prune.in
Ignored: references/chr22.filt.prune.out
Ignored: references/chr22.filtered.log
Ignored: references/chr22.filtered.nosex
Ignored: references/chr22.filtered.vcf
Ignored: references/chr22.filtered.vcf.bed
Ignored: references/chr22.filtered.vcf.bim
Ignored: references/chr22.filtered.vcf.fam
Ignored: references/chr22.filtered.vcf.log
Ignored: references/chr22.filtered.vcf.nosex
Ignored: references/chr22.filtered.vcf.rel
Ignored: references/chr22.filtered.vcf.rel.id
Ignored: references/chr22.filtered.vcf.rel_mod
Ignored: references/chr22.genes.gff3
Ignored: references/chr22.genes.gff3_annotation
Ignored: references/chr22.genes.gff3_chunks
Ignored: references/chr22.vcf.gz
Ignored: references/eqtl_summary_stats.tar.gz
Ignored: references/eqtl_summary_stats_renamed/
Ignored: references/keep_samples.txt
Ignored: references/remove_snps.txt
Ignored: references/test.genome
Ignored: references/test.log
Ignored: references/test.nosex
Untracked files:
Untracked: .DS_Store
Untracked: .cache/
Untracked: .config/
Untracked: .snakemake/
Untracked: KEJP_2020_splatPop.Rproj
Untracked: analysis/bug_fixing.Rmd
Untracked: code/splatter_1.17.2.tar.gz
Untracked: docs/.DS_Store
Unstaged changes:
Modified: .gitignore
Modified: .gitlab-ci.yml
Modified: CITATION
Modified: Dockerfile
Modified: LICENSE
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/10x-Fibrob_simulations.Rmd
Modified: analysis/10x-Neuro_estimate-params.Rmd
Modified: analysis/10x-Neuro_simulations.Rmd
Modified: analysis/KEJP_iPSC-ss2.Rmd
Modified: analysis/_site.yml
Modified: analysis/about.Rmd
Modified: analysis/index.Rmd
Modified: analysis/license.Rmd
Modified: analysis/ss2-iPSC_estimate-params.Rmd
Modified: analysis/ss2-iPSC_simulations.Rmd
Modified: cluster.json
Modified: code/1_process-empirical-data.R
Modified: code/2_estimate-splatPopParams.R
Modified: code/4_simulate_DEG.R
Modified: code/README.md
Modified: code/plot_functions.R
Modified: code/plot_functions2.R
Modified: data/README.md
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.css
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.js
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cerulean.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cosmo.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/darkly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/flatly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Lato.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycle.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycleBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSans.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBoldItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLightItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Raleway.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RalewayBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Roboto.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoMedium.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansPro.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Ubuntu.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/journal.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/lumen.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/paper.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/readable.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/sandstone.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/simplex.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/spacelab.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/united.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/yeti.min.css
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.eot
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.svg
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.ttf
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff2
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.js
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.min.js
Modified: docs/site_libs/bootstrap-3.3.5/js/npm.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/html5shiv.min.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/respond.min.js
Modified: docs/site_libs/font-awesome-5.1.0/css/all.css
Modified: docs/site_libs/font-awesome-5.1.0/css/v4-shims.css
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff2
Modified: docs/site_libs/header-attrs-2.5/header-attrs.js
Modified: docs/site_libs/jquery-1.11.3/jquery.min.js
Modified: docs/site_libs/jqueryui-1.11.4/README
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_444444_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_555555_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777620_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777777_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_cc0000_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_ffffff_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/index.html
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.min.css
Modified: docs/site_libs/navigation-1.1/codefolding.js
Modified: docs/site_libs/navigation-1.1/sourceembed.js
Modified: docs/site_libs/navigation-1.1/tabsets.js
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.css
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.js
Modified: environment.yml
Modified: envs/limix_env.yaml
Modified: envs/myenv.yaml
Modified: org/README.md
Modified: org/project_management.org
Modified: output/README.md
Modified: references/README.md
Modified: resources/README.md
Modified: resources/keep_samples.txt
Modified: workflow/config_bp_ss2.yaml
Modified: workflow/sims_limix_v1.15.1.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ss2-iPSC_simulations.Rmd) and HTML (public/ss2-iPSC_simulations.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 116e4a3 | cazodi | 2021-10-08 | update mean-variance plots for manuscript |
| html | 116e4a3 | cazodi | 2021-10-08 | update mean-variance plots for manuscript |
| Rmd | bd0c8b0 | cazodi | 2021-05-17 | updates to 10x neuroseq and ss2 ipsc examples |
| html | bd0c8b0 | cazodi | 2021-05-17 | updates to 10x neuroseq and ss2 ipsc examples |
| Rmd | b1e4853 | cazodi | 2021-03-16 | update plot functions |
| Rmd | ae3af3e | cazodi | 2021-03-10 | update simple ss2 ipsc sim |
| Rmd | d24c101 | cazodi | 2021-02-17 | ipsc ss2 preprocessing |
#install.packages("/mnt/mcscratch/cazodi/Software/splatter_1.15.2.tar.gz", repos = NULL, type="source")
suppressPackageStartupMessages({
library(SingleCellExperiment)
library(scater)
library(tidyverse)
#detach("package:splatter", unload=TRUE)
library(splatter)
library(VariantAnnotation)
library(cluster)
library(fitdistrplus)
library(RColorBrewer)
library(ggpubr)
library(cowplot)
})
source("code/plot_functions.R")
source("code/misc_functions.R")date <- Sys.Date()
set.seed(42)
n.genes <- 504
save <- TRUE
rerun <- FALSE
date.use <- "2021-05-14"
sample.colors <- projectColors("samples")
# Chromosome 22 data
gff <- read.table("references/chr22.genes.gff3", sep="\t", header=FALSE, quote="")
vcf <- readVcf("references/chr22.filtered.vcf", "hg38")
sampleNames <- colnames(geno(vcf)$GT)
# Smartseq2 iPSC data and splatPopParams
sce <- readRDS("data/sce_iPSC-ss2_D0.rds")
params <- readRDS("output/01_sims/splatPop-params_iPSC-ss2_sc.rds")
params.pb <- readRDS("output/01_sims/splatPop-params_iPSC-ss2_psudoB.rds")Simple simulations
Cell-level visualization
- PCA plots: relative relationship between cells from different individuals, batches, cell-groups, etc.
- Clustering statistics: quantify the degree of separation between cells from the same compared to different clusters, where the cluster refers to the donor, batch, cell-group, or cohort. Note that the Silhouette width is based on the compactness vs. separation from the nearest neighbor cluster.
Gene-level visualizations
- Percent of variance explained: Percent of variance in expression across cells explained by each experimental factor (e.g., individual, batch, cell-group).
- Mean-variance relationship: The mean-variance trend for each gene across, with counts mean aggregated by individual (or individual-batch, etc), using a range of nCells per donor from the simulated data.
sce.simple <- subset(sce, , Batch == "expt_37")
if(rerun){
nCells <- as.data.frame(colData(sce.simple)) %>%
group_by(Sample) %>% dplyr::count()
nCells <- round(mean(nCells$n))
nSamples <- length(unique(sce.simple$Sample))
vcf.simple <- vcf[, sample(sampleNames, nSamples)]
params.simple <- setParams(params,
eqtl.n = 0.75,
similarity.scale = 2,
batchCells = c(nCells))
sim.simple <- splatPopSimulate(vcf = vcf.simple,
gff = gff,
params = params.simple,
sparsify = FALSE)
}else{
sim.simple <- readRDS(paste0("output/01_sims/", date.use, "_simple-ss2.rds"))
}
source("code/plot_functions.R")
plotComparisons(sim=sim.simple, emp=sce.simple, maxCells=50,
mv.nCells = c(5, 20, 52), mv.y.max = 0.8) `geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.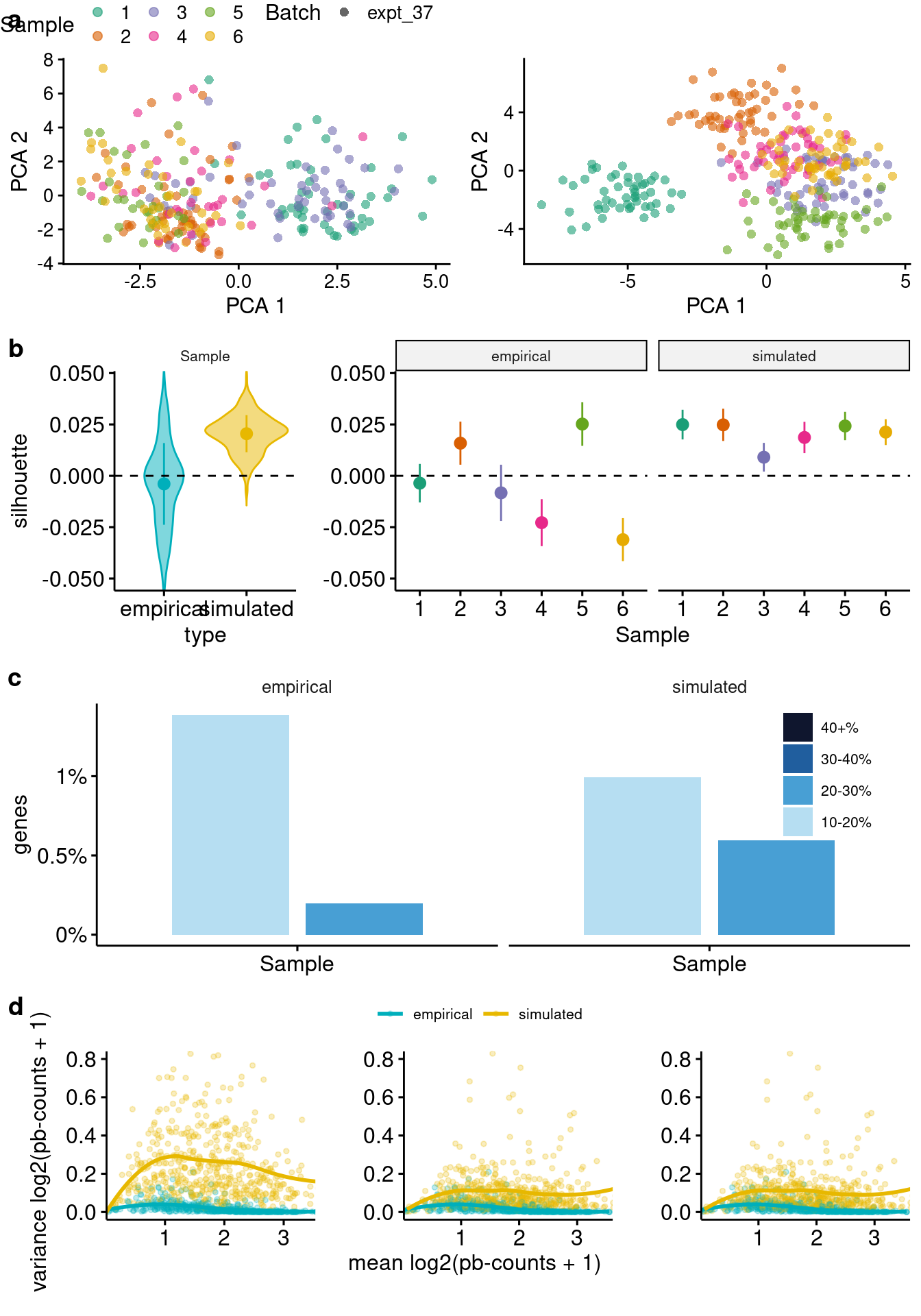
if(save){
save.name <- paste0(date, "_simple-ss2")
saveRDS(sim.simple, paste0("output/01_sims/", save.name, ".rds"))
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 6, height = 7)
}Simulations from “bulk” reference data.
While the single-cell parameters for splatPop need to be estimated from real single-cell data, the population scale parameters can be estimated from single-cell or bulk population scale RNA-seq data. To ensure the simulated data reflects single-cell data, if bulk data is used to estimate population parameters, splatPop will perform a quantile normalization step, where for each sample, the simulated gene means are quantile normalized to match the distribution of the gene means from the single-cell data. Note that this step can change the mean-variance relationship of the simulated data, so care should be taken when simulating data from bulk estimated parameters.
if(rerun){
params.pb <- setParams(params.pb,
eqtl.n = 0.75,
similarity.scale = 4,
batchCells = c(nCells))
sim.ss2.pb <- splatPopSimulate(vcf = vcf.simple, gff = gff,
params = params.pb, sparsify = FALSE)
}else{
sim.ss2.pb <- readRDS(paste0("output/01_sims/", date.use, "_simple-ss2-pb.rds"))
}
plotComparisons(sim=sim.ss2.pb, emp=sce.simple, maxCells=50,
mv.nCells = c(5, 20, 52), mv.y.max = 0.8)`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.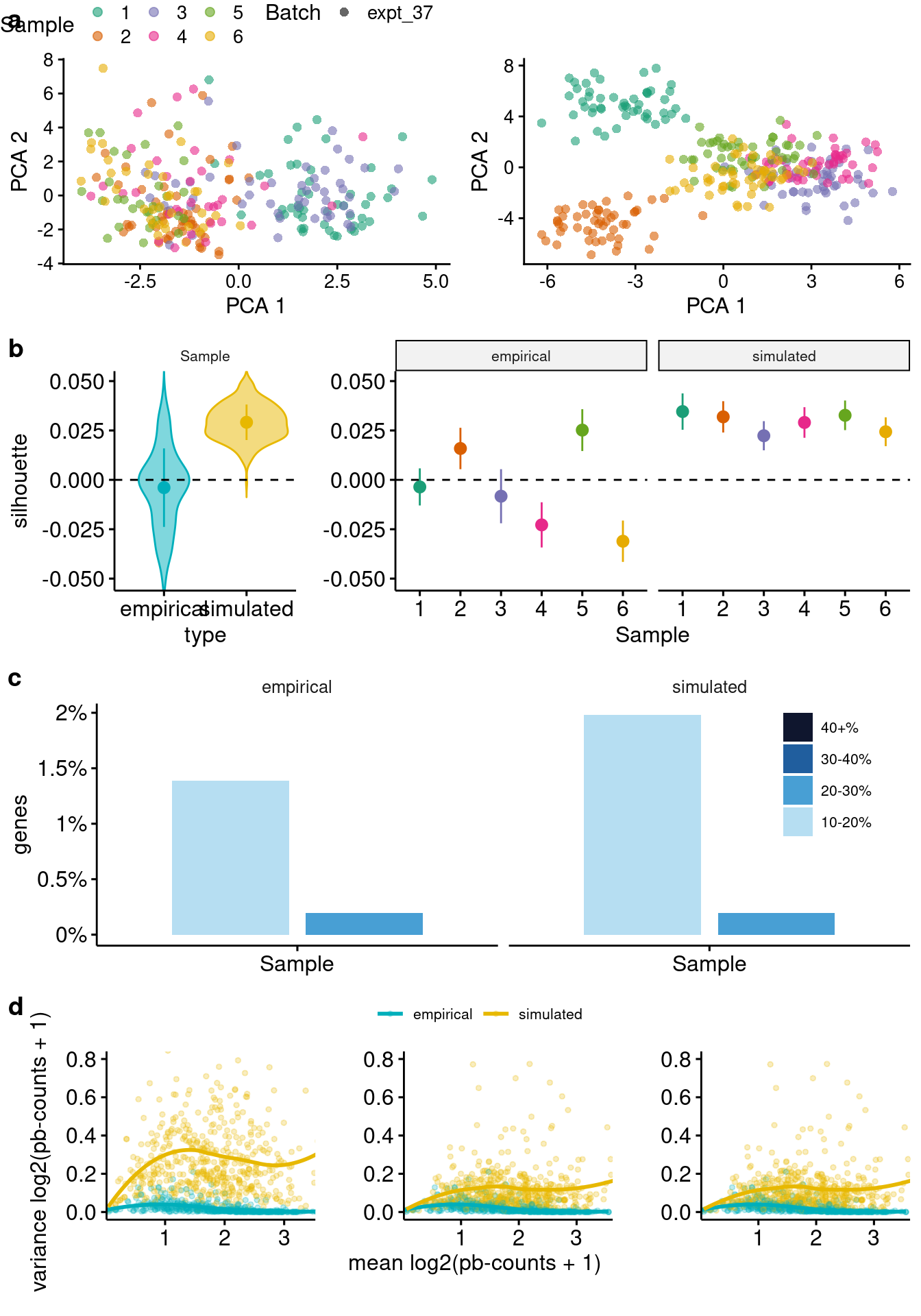
if(save){
save.name <- paste0(date, "_simple-ss2-pb")
saveRDS(sim.ss2.pb, paste0("output/01_sims/", save.name, ".rds"))
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 6, height = 7)
}Batch effects
Simple example
vcf.batches.simple <- vcf[, sample(sampleNames, 5)]
if(rerun){
params.ss.batches.simple <- setParams(params, batchCells=c(40, 40),
batch.size = 3,
eqtl.n = 0.5,
batch.facLoc = 0.1,
batch.facScale = 0.25)
sim.batches.simple <- splatPopSimulate(vcf = vcf.batches.simple, gff = gff,
params = params.ss.batches.simple,
sparsify = FALSE)
}else{
sim.batches.simple <- readRDS(paste0("output/01_sims/", date.use,
"_example-batches.rds"))
}
plotSims(sim=sim.batches.simple, variables = c("Sample", "Batch"),
colour_by = "Sample", shape_by= "Batch")Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.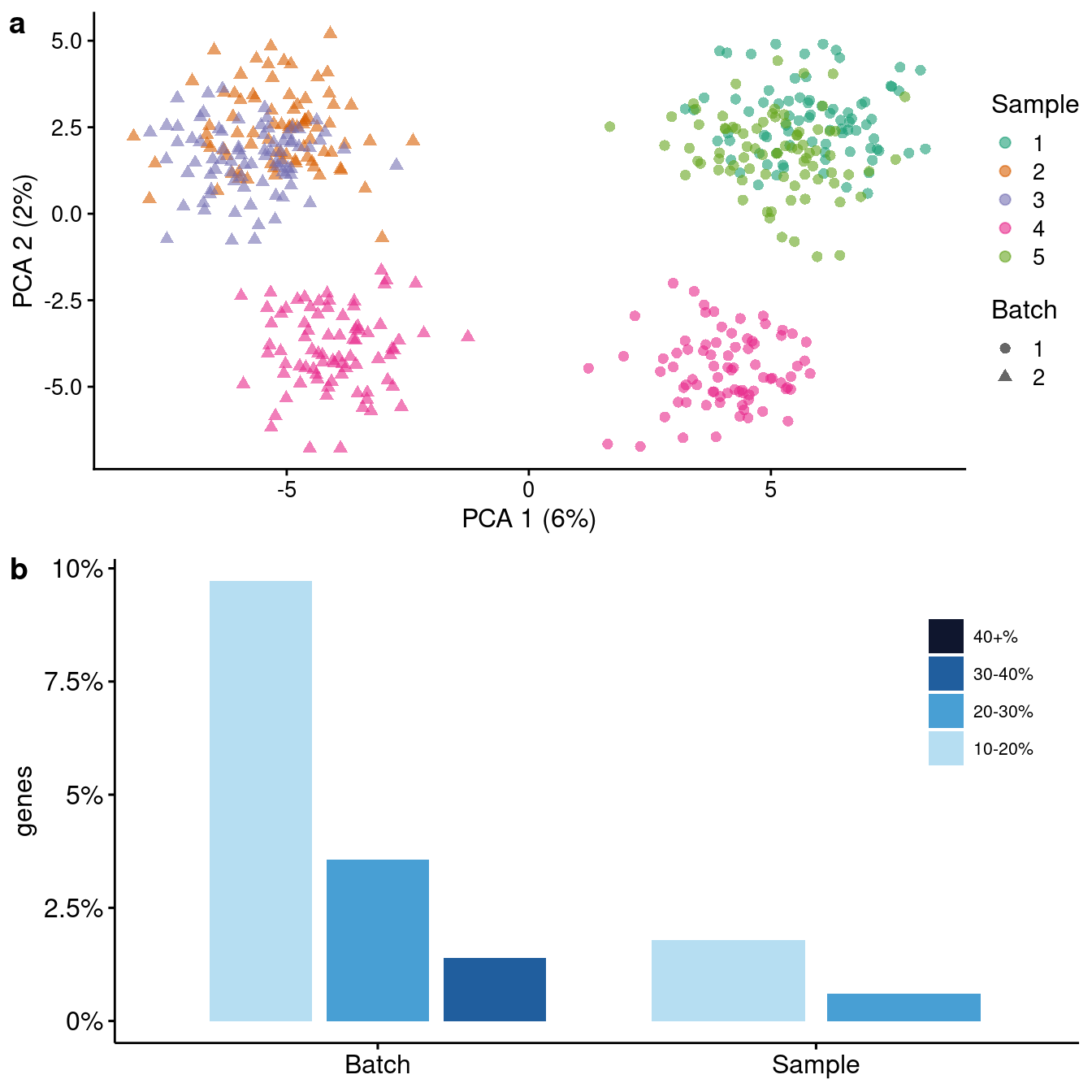
if(save){
save.name <- paste0(date, "_example-batches")
saveRDS(sim.batches.simple, paste0("output/01_sims/", save.name, ".rds"))
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 3, height = 5)
}Smartseq2 - iPSC complex batch structure
The ss2-iPSC data set contains data from 10 individuals sequenced over 3 batches (expt 31, 32, and 33), with two individuals () replicated in two batches.
sce.batches <- subset(sce, , Batch %in% c("expt_22", "expt_23", "expt_24"))
#sce.batches <- subset(sce, , Batch %in% c("expt_37", "expt_38", "expt_39"))
sce.batches <- subset(sce.batches, , !(Sample %in% c("eoxi", "iudw", "oikd")))
if(rerun){
table(sce.batches$Sample, sce.batches$Batch)
nSamples <- length(unique(sce.batches$Sample))
nCells.SB <- as.list(data.frame(table(sce.batches$Sample,
sce.batches$Batch)))$Freq
nCells.SB <- nCells.SB[which(nCells.SB != 0)]
nC.fit <- fitdist(nCells.SB, "gamma")
vcf.batches <- vcf[, sample(sampleNames, nSamples)]
params.ss.batches <- setParams(params,
# population description parameters
nCells.sample = TRUE,
nCells.shape = nC.fit$estimate["shape"],
nCells.rate = nC.fit$estimate["rate"],
batchCells=c(1, 1, 1),
batch.size = 4,
# parameters specifying effects
eqtl.n = 1,
similarity.scale = 7,
batch.facLoc = c(0, 0, 0),
batch.facScale = c(0.35, 0.01, 0.01))
sim.batches <- splatPopSimulate(vcf = vcf.batches, gff = gff,
params = params.ss.batches, sparsify = FALSE)
} else{
sim.batches <- readRDS(paste0("output/01_sims/", date.use, "_batches.rds"))
}
plotComparisons(sim=sim.batches, emp=sce.batches,
maxCells = 30, maxBatches=3,
variables = c("Sample", "Batch"),
colour_by = "Sample",
shape_by= "Batch",
samp.col = sample.colors,
mv.nCells = c(5, 20, 52),
mv.y.max = 0.8)`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'
`geom_smooth()` using formula 'y ~ x'Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.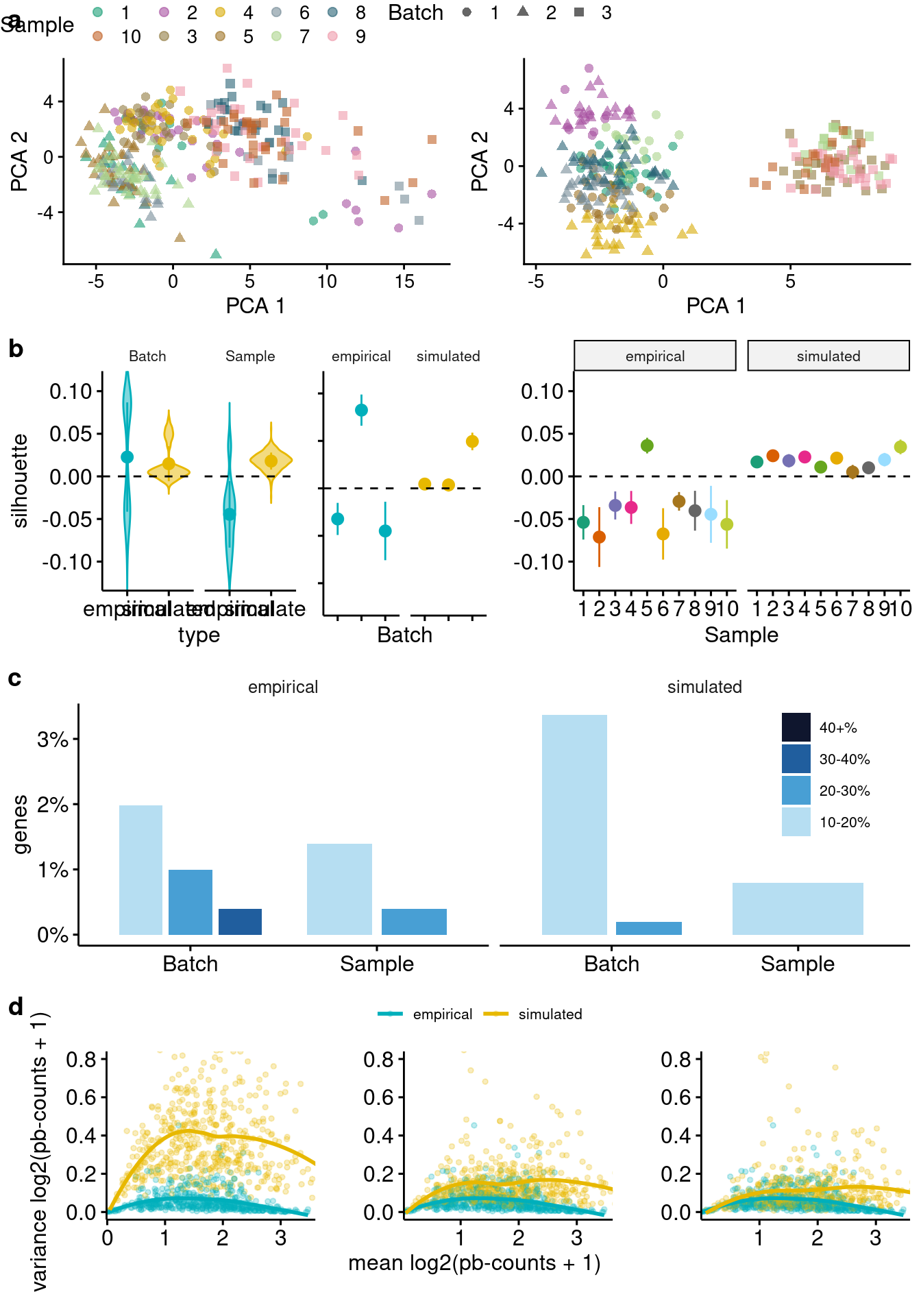
if(save){
save.name <- paste0(date, "_batches")
saveRDS(sim.batches, paste0("output/01_sims/", save.name, ".rds"))
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 6, height = 7)
}tSNE plot comparisons
While the PCA plots are useful for showing global relationships between cells, UMAP and tSNE dimension reduction methods have become the norm for visualizing single-cell RNA-sequencing data.
pSim <- plotTSNEx(sim.simple, colour_by="Sample",
maxCells = 50, samp.col = sample.colors)Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.pEmp<- plotTSNEx(sce.simple, colour_by="Sample",
maxCells = 50, samp.col = sample.colors) Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.pSimBatch <- plotTSNEx(sim.batches, colour_by="Sample", shape_by="Batch",
maxCells = 50, samp.col = sample.colors)Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.pEmpBatch <- plotTSNEx(sce.batches, colour_by="Sample", shape_by="Batch",
maxCells = 50, samp.col = sample.colors) Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.plot_grid(pEmp, pSim, pEmpBatch, pSimBatch, ncol=2, labels = "auto")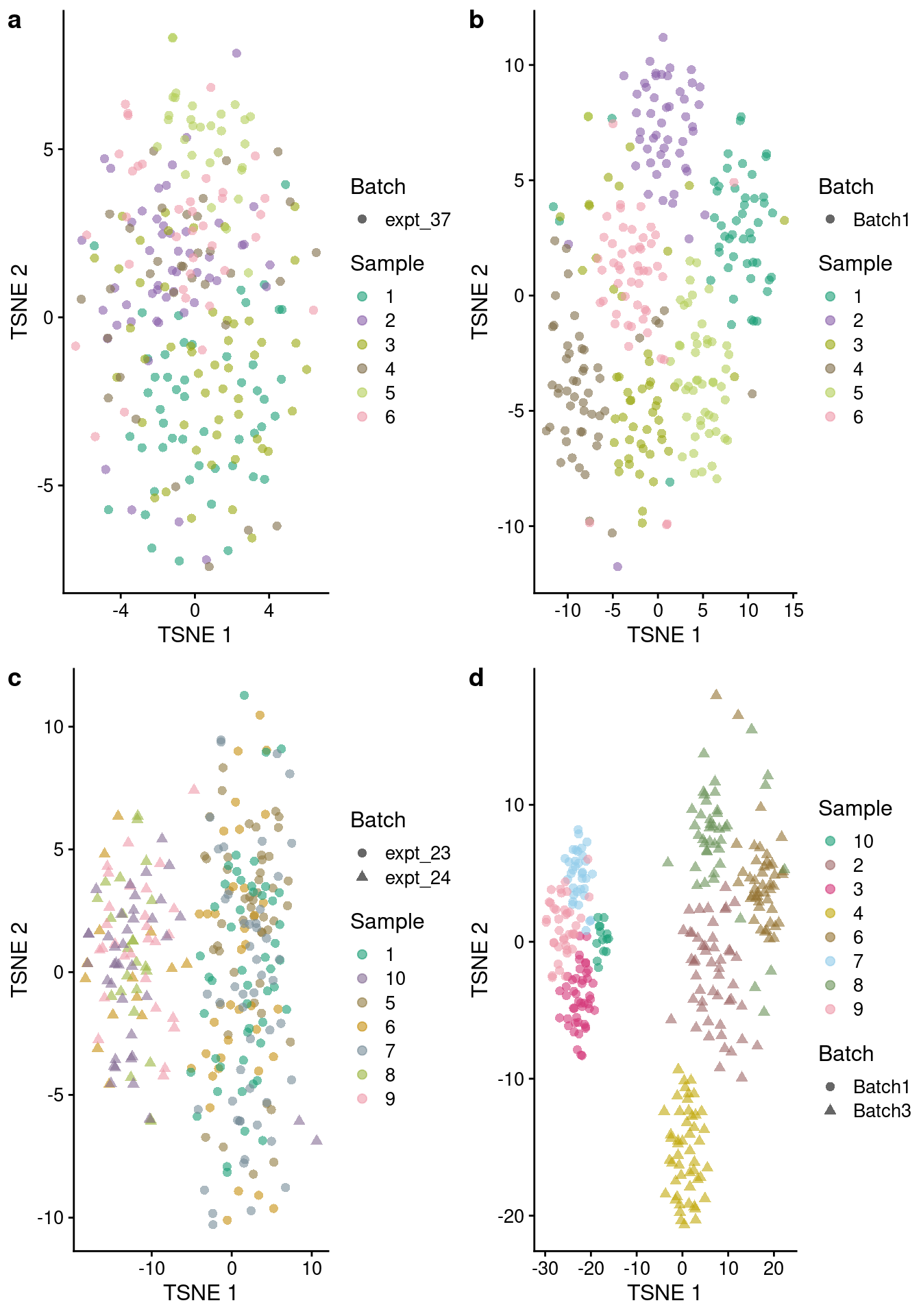
if(save){
save.name <- paste0(date, "_ss2-TSNEs")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 6)
}Saving 6 x 10 in imageLarge simulation for eQTL mapping demo
Using the same parameters as above, we will simulate a larger dataset to perform eQTL mapping on. We will simulate 500 cells per sample, with samples sequenced in 10 batches, with 10 samples per batch (total = 100 samples). We randomly sample the batch effect sizes from normal distributions around the values used above.
if(rerun) {
nSamples <- 100
vcf.large <- vcf[, sample(sampleNames, nSamples)]
params.ss.large <- setParams(params,
# population description parameters
nCells.sample = FALSE,
batchCells= rep(500, 10),
batch.size = 10,
# parameters specifying effects
eqtl.n = 0.7,
similarity.scale = 7,
batch.facLoc = abs(rnorm(10, 0.05, 0.1)),
batch.facScale = abs(rnorm(10, 0.15, 0.1)))
sim.large <- splatPopSimulate(vcf = vcf.large, gff = gff,
params = params.ss.large, sparsify = FALSE)
if(save){
save.name <- paste0(date, "_ss2-large")
saveRDS(sim.large, paste0("output/01_sims/", save.name, ".rds"))
}
}
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.0.4 (2021-02-15)
os Red Hat Enterprise Linux 8.4 (Ootpa)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-11-15
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.0.2)
AnnotationDbi 1.52.0 2020-10-27 [1] Bioconductor
askpass 1.1 2019-01-13 [1] CRAN (R 4.0.2)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.2)
backports 1.2.1 2020-12-09 [1] CRAN (R 4.0.4)
beachmat 2.6.4 2020-12-20 [1] Bioconductor
beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.0.4)
Biobase * 2.50.0 2020-10-27 [1] Bioconductor
BiocFileCache 1.14.0 2020-10-27 [1] Bioconductor
BiocGenerics * 0.36.1 2021-04-16 [1] Bioconductor
BiocNeighbors 1.8.2 2020-12-07 [1] Bioconductor
BiocParallel 1.24.1 2020-11-06 [1] Bioconductor
BiocSingular 1.6.0 2020-10-27 [1] Bioconductor
biomaRt 2.46.3 2021-02-09 [1] Bioconductor
Biostrings * 2.58.0 2020-10-27 [1] Bioconductor
bit 4.0.4 2020-08-04 [1] CRAN (R 4.0.2)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.0.2)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.0.4)
blob 1.2.2 2021-07-23 [1] CRAN (R 4.0.4)
broom 0.7.9 2021-07-27 [1] CRAN (R 4.0.4)
BSgenome 1.58.0 2020-10-27 [1] Bioconductor
bslib 0.2.5.1 2021-05-18 [1] CRAN (R 4.0.4)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.4)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.0.4)
car 3.0-11 2021-06-27 [1] CRAN (R 4.0.4)
carData 3.0-4 2020-05-22 [1] CRAN (R 4.0.2)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.0.2)
checkmate 2.0.0 2020-02-06 [1] CRAN (R 4.0.2)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.0.4)
cluster * 2.1.0 2019-06-19 [2] CRAN (R 4.0.4)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.0.4)
cowplot * 1.1.1 2020-12-30 [1] CRAN (R 4.0.4)
crayon 1.4.1 2021-02-08 [1] CRAN (R 4.0.4)
curl 4.3.2 2021-06-23 [1] CRAN (R 4.0.4)
data.table 1.14.2 2021-09-27 [1] CRAN (R 4.0.4)
DBI 1.1.1 2021-01-15 [1] CRAN (R 4.0.4)
dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.0.4)
DelayedArray 0.16.3 2021-03-24 [1] Bioconductor
DelayedMatrixStats 1.12.3 2021-02-03 [1] Bioconductor
desc 1.4.0 2021-09-28 [1] CRAN (R 4.0.4)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.0.4)
digest 0.6.28 2021-09-23 [1] CRAN (R 4.0.4)
dplyr * 1.0.7 2021-06-18 [1] CRAN (R 4.0.4)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.0.4)
evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.2)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.0.4)
farver 2.1.0 2021-02-28 [1] CRAN (R 4.0.4)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.0.3)
fitdistrplus * 1.1-6 2021-09-28 [1] CRAN (R 4.0.4)
forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.0.4)
foreign 0.8-81 2020-12-22 [2] CRAN (R 4.0.4)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
GenomeInfoDb * 1.26.7 2021-04-08 [1] Bioconductor
GenomeInfoDbData 1.2.4 2020-11-10 [1] Bioconductor
GenomicAlignments 1.26.0 2020-10-27 [1] Bioconductor
GenomicFeatures 1.42.3 2021-04-01 [1] Bioconductor
GenomicRanges * 1.42.0 2020-10-27 [1] Bioconductor
ggbeeswarm 0.6.0 2017-08-07 [1] CRAN (R 4.0.2)
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.0.4)
ggpubr * 0.4.0 2020-06-27 [1] CRAN (R 4.0.3)
ggsignif 0.6.3 2021-09-09 [1] CRAN (R 4.0.4)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.0.4)
glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.0.2)
gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.2)
haven 2.4.3 2021-08-04 [1] CRAN (R 4.0.4)
highr 0.9 2021-04-16 [1] CRAN (R 4.0.4)
hms 1.1.0 2021-05-17 [1] CRAN (R 4.0.4)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.0.4)
httpuv 1.6.3 2021-09-09 [1] CRAN (R 4.0.4)
httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
IRanges * 2.24.1 2020-12-12 [1] Bioconductor
irlba 2.3.3 2019-02-05 [1] CRAN (R 4.0.2)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.4)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.4)
knitr 1.34 2021-09-09 [1] CRAN (R 4.0.4)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.0.2)
later 1.3.0 2021-08-18 [1] CRAN (R 4.0.4)
lattice 0.20-41 2020-04-02 [2] CRAN (R 4.0.4)
lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.0.4)
locfit 1.5-9.4 2020-03-25 [1] CRAN (R 4.0.2)
lubridate 1.8.0 2021-10-07 [1] CRAN (R 4.0.4)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.3)
MASS * 7.3-54 2021-05-03 [1] CRAN (R 4.0.4)
Matrix 1.3-4 2021-06-01 [1] CRAN (R 4.0.4)
MatrixGenerics * 1.2.1 2021-01-30 [1] Bioconductor
matrixStats * 0.60.0 2021-07-26 [1] CRAN (R 4.0.4)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.4)
mgcv 1.8-36 2021-06-01 [1] CRAN (R 4.0.4)
modelr 0.1.8 2020-05-19 [1] CRAN (R 4.0.2)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.2)
nlme 3.1-153 2021-09-07 [1] CRAN (R 4.0.4)
openssl 1.4.5 2021-09-02 [1] CRAN (R 4.0.4)
openxlsx 4.2.4 2021-06-16 [1] CRAN (R 4.0.4)
pillar 1.6.3 2021-09-26 [1] CRAN (R 4.0.4)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.4)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
pkgload 1.2.2 2021-09-11 [1] CRAN (R 4.0.4)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.2)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.0.4)
progress 1.2.2 2019-05-16 [1] CRAN (R 4.0.2)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.0.4)
ps 1.6.0 2021-02-28 [1] CRAN (R 4.0.4)
purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.0.4)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.0.4)
RColorBrewer * 1.1-2 2014-12-07 [1] CRAN (R 4.0.2)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.0.4)
RCurl 1.98-1.4 2021-08-17 [1] CRAN (R 4.0.4)
readr * 2.0.1 2021-08-10 [1] CRAN (R 4.0.4)
readxl 1.3.1 2019-03-13 [1] CRAN (R 4.0.2)
remotes 2.4.1 2021-09-29 [1] CRAN (R 4.0.4)
reprex 2.0.1 2021-08-05 [1] CRAN (R 4.0.4)
rio 0.5.27 2021-06-21 [1] CRAN (R 4.0.4)
rlang 0.4.12 2021-10-18 [1] CRAN (R 4.0.4)
rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.0.4)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.3)
Rsamtools * 2.6.0 2020-10-27 [1] Bioconductor
RSQLite 2.2.8 2021-08-21 [1] CRAN (R 4.0.4)
rstatix 0.7.0 2021-02-13 [1] CRAN (R 4.0.4)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.3)
rsvd 1.0.5 2021-04-16 [1] CRAN (R 4.0.4)
rtracklayer 1.50.0 2020-10-27 [1] Bioconductor
Rtsne 0.15 2018-11-10 [1] CRAN (R 4.0.4)
rvest 1.0.1 2021-07-26 [1] CRAN (R 4.0.4)
S4Vectors * 0.28.1 2020-12-09 [1] Bioconductor
sass 0.4.0 2021-05-12 [1] CRAN (R 4.0.4)
scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.2)
scater * 1.18.6 2021-02-26 [1] Bioconductor
scuttle 1.0.4 2020-12-17 [1] Bioconductor
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
SingleCellExperiment * 1.12.0 2020-10-27 [1] Bioconductor
sparseMatrixStats 1.2.1 2021-02-02 [1] Bioconductor
splatter * 1.14.1 2020-12-01 [1] Bioconductor
stringi 1.7.5 2021-10-04 [1] CRAN (R 4.0.4)
stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
SummarizedExperiment * 1.20.0 2020-10-27 [1] Bioconductor
survival * 3.2-13 2021-08-24 [1] CRAN (R 4.0.4)
testthat 3.0.4 2021-07-01 [1] CRAN (R 4.0.4)
tibble * 3.1.4 2021-08-25 [1] CRAN (R 4.0.4)
tidyr * 1.1.4 2021-09-27 [1] CRAN (R 4.0.4)
tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.0.4)
tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 4.0.4)
tzdb 0.1.2 2021-07-20 [1] CRAN (R 4.0.4)
usethis 2.0.1 2021-02-10 [1] CRAN (R 4.0.4)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.4)
VariantAnnotation * 1.36.0 2020-10-27 [1] Bioconductor
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.0.4)
vipor 0.4.5 2017-03-22 [1] CRAN (R 4.0.2)
viridis 0.6.2 2021-10-13 [1] CRAN (R 4.0.4)
viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.0.4)
whisker 0.4 2019-08-28 [1] CRAN (R 4.0.2)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.0.4)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.0.2)
xfun 0.26 2021-09-14 [1] CRAN (R 4.0.4)
XML 3.99-0.7 2021-08-17 [1] CRAN (R 4.0.4)
xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.2)
XVector * 0.30.0 2020-10-27 [1] Bioconductor
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
zip 2.2.0 2021-05-31 [1] CRAN (R 4.0.4)
zlibbioc 1.36.0 2020-10-27 [1] Bioconductor
[1] /mnt/mcfiles/cazodi/R/x86_64-pc-linux-gnu-library/4.0
[2] /opt/R/4.0.4/lib/R/library