Case study: t-test with only zeros
Last updated: 2024-01-01
Checks: 7 0
Knit directory:
mage_2020_marker-gene-benchmarking/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190102) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2c5ed16. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Renviron
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .snakemake/
Ignored: NSForest/.Rhistory
Ignored: NSForest/NS-Forest_v3_Extended_Binary_Markers_Supplmental.csv
Ignored: NSForest/NS-Forest_v3_Full_Results.csv
Ignored: NSForest/NSForest3_medianValues.csv
Ignored: NSForest/NSForest_v3_Final_Result.csv
Ignored: NSForest/__pycache__/
Ignored: NSForest/data/
Ignored: RankCorr/picturedRocks/__pycache__/
Ignored: benchmarks/
Ignored: config/
Ignored: data/cellmarker/
Ignored: data/downloaded_data/
Ignored: data/expert_annotations/
Ignored: data/expert_mgs/
Ignored: data/raw_data/
Ignored: data/real_data/
Ignored: data/sim_data/
Ignored: data/sim_mgs/
Ignored: data/special_real_data/
Ignored: figures/
Ignored: logs/
Ignored: results/
Ignored: weights/
Unstaged changes:
Modified: smash-fork
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/t-test-only-zero-case-study.Rmd) and HTML
(public/t-test-only-zero-case-study.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2c5ed16 | Jeffrey Pullin | 2024-01-01 | Tweak website |
| html | 39c3c99 | Jeffrey Pullin | 2024-01-01 | Remove obselete files in public/ |
| Rmd | 35e497e | Jeffrey Pullin | 2023-11-21 | Remove unneeded analysis files |
| html | fcecf65 | Jeffrey Pullin | 2022-09-09 | Build site. |
| html | af96b34 | Jeffrey Pullin | 2022-08-30 | Build site. |
| html | 0e47874 | Jeffrey Pullin | 2022-05-04 | Build site. |
| html | b5045c1 | Jeffrey Pullin | 2022-05-02 | Build site. |
| Rmd | 048156f | Jeffrey Pullin | 2022-05-02 | Tweak analysis outputs |
| html | 048156f | Jeffrey Pullin | 2022-05-02 | Tweak analysis outputs |
| html | 8b989e1 | Jeffrey Pullin | 2022-05-02 | Build site. |
| html | 0548273 | Jeffrey Pullin | 2022-05-02 | Build site. |
| Rmd | 50bca7c | Jeffrey Pullin | 2022-05-02 | workflowr::wflow_publish(all = TRUE, republish = TRUE) |
| html | 50bca7c | Jeffrey Pullin | 2022-05-02 | workflowr::wflow_publish(all = TRUE, republish = TRUE) |
| html | 5cc008f | Jeffrey Pullin | 2022-02-09 | Build site. |
| Rmd | d1aca16 | Jeffrey Pullin | 2022-02-09 | Refresh website |
| Rmd | aca9ad2 | Jeffrey Pullin | 2021-11-29 | Various changes made in the last days before thesis submission |
| Rmd | d3804d1 | Jeffrey Pullin | 2021-09-23 | Polish analyses |
| Rmd | 17f2a0f | Jeffrey Pullin | 2021-08-07 | Add new plots and analysis for lab meeting 5/8/2021 |
| Rmd | 218574a | Jeffrey Pullin | 2021-07-23 | Add (rough) draft of t-test zero case study |
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(ggplot2)
library(purrr)
library(patchwork)
library(tibble)
library(SingleCellExperiment)Loading required package: SummarizedExperimentLoading required package: MatrixGenericsLoading required package: matrixStats
Attaching package: 'matrixStats'The following object is masked from 'package:dplyr':
count
Attaching package: 'MatrixGenerics'The following objects are masked from 'package:matrixStats':
colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
colWeightedMeans, colWeightedMedians, colWeightedSds,
colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
rowWeightedSds, rowWeightedVarsLoading required package: GenomicRangesLoading required package: stats4Loading required package: BiocGenerics
Attaching package: 'BiocGenerics'The following objects are masked from 'package:dplyr':
combine, intersect, setdiff, unionThe following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, append, as.data.frame, basename, cbind, colnames,
dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
union, unique, unsplit, which.max, which.minLoading required package: S4Vectors
Attaching package: 'S4Vectors'The following objects are masked from 'package:dplyr':
first, renameThe following objects are masked from 'package:base':
expand.grid, I, unnameLoading required package: IRanges
Attaching package: 'IRanges'The following object is masked from 'package:purrr':
reduceThe following objects are masked from 'package:dplyr':
collapse, desc, sliceLoading required package: GenomeInfoDbLoading required package: BiobaseWelcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
Attaching package: 'Biobase'The following object is masked from 'package:MatrixGenerics':
rowMediansThe following objects are masked from 'package:matrixStats':
anyMissing, rowMedianspbmc3k <- readRDS(here::here("data", "real_data", "pbmc3k.rds"))
endothelial <- readRDS(here::here("data", "real_data", "endothelial.rds"))
lawlor <- readRDS(here::here("data", "real_data", "lawlor.rds"))
mgs_t_raw <- readRDS(
here::here("results", "real_data", "pbmc3k-seurat_t.rds")
)mgs_t_platelet <- mgs_t_raw %>%
pluck("result") %>%
filter(cluster == "Platelet")
count_platelet_data <- data.frame(
gene = rownames(pbmc3k),
count = rowSums(counts(pbmc3k)[, colLabels(pbmc3k) == "Platelet"])
)
mgs_t_platelet <- left_join(mgs_t_platelet, count_platelet_data, by = "gene")mgs_t_platelet %>%
print(n = 300)# A tibble: 768 × 8
p_value p_value_adj cluster log_fc gene raw_statistic scaled_statistic
<dbl> <dbl> <fct> <dbl> <chr> <dbl> <dbl>
1 0 0 Platelet -2.24 UBXN1 0 0
2 0 0 Platelet -2.35 EVL 0 0
3 0 0 Platelet -2.90 ZFP3… 0 0
4 0 0 Platelet -2.92 CD3D 0 0
5 Inf. e-324 1.98e-320 Platelet -1.99 HNRN… 0 0
6 3.84e-277 7.69e-274 Platelet -1.84 EIF3D 0 0
7 1.67e-268 3.34e-265 Platelet -2.26 IFI6 0 0
8 3.74e-244 7.49e-241 Platelet -1.78 LCK 0 0
9 2.00e-243 4.01e-240 Platelet -2.07 CD7 0 0
10 3.18e-229 6.36e-226 Platelet -1.56 CD164 0 0
11 5.00e-225 1.00e-221 Platelet -1.61 CSNK… 0 0
12 4.13e-218 8.26e-215 Platelet -1.58 SRP9 0 0
13 1.15e-216 2.31e-213 Platelet -1.52 RSL1… 0 0
14 5.15e-214 1.03e-210 Platelet -1.49 ATP5… 0 0
15 1.02e-205 2.03e-202 Platelet -1.48 SON 0 0
16 1.94e-205 3.88e-202 Platelet -1.77 CD2 0 0
17 3.99e-204 7.97e-201 Platelet -1.48 RBMX 0 0
18 2.70e-196 5.39e-193 Platelet -1.47 TBC1… 0 0
19 5.80e-196 1.16e-192 Platelet -1.56 ACAP1 0 0
20 3.35e-190 6.70e-187 Platelet -1.44 GYPC 0 0
21 1.11e-189 2.22e-186 Platelet -1.37 UQCRQ 0 0
22 7.29e-188 1.46e-184 Platelet -1.46 DNAJ… 0 0
23 1.80e-186 3.60e-183 Platelet -1.48 TMBI… 0 0
24 6.64e-182 1.33e-178 Platelet -1.49 XBP1 0 0
25 6.42e-180 1.28e-176 Platelet -1.63 GIMA… 0 0
26 2.54e-178 5.07e-175 Platelet -1.36 ARHG… 0 0
27 4.70e-176 9.40e-173 Platelet -1.35 NCOR1 0 0
28 1.96e-174 3.93e-171 Platelet -1.46 RPL39 0 0
29 2.98e-174 5.96e-171 Platelet -1.39 CASP1 0 0
30 3.04e-174 6.08e-171 Platelet -1.64 RGS2 0 0
31 1.04e-165 2.09e-162 Platelet -1.62 CD247 0 0
32 3.56e-164 7.11e-161 Platelet -1.29 LMAN2 0 0
33 3.11e-161 6.22e-158 Platelet -1.47 DUSP2 0 0
34 2.34e-160 4.69e-157 Platelet -1.31 PIK3… 0 0
35 5.18e-159 1.04e-155 Platelet -1.32 RHOH 0 0
36 1.60e-158 3.21e-155 Platelet -1.30 BIN1 0 0
37 5.83e-155 1.17e-151 Platelet -1.21 EEF1G 0 0
38 7.21e-154 1.44e-150 Platelet -1.19 KHDR… 0 0
39 6.67e-152 1.33e-148 Platelet -1.22 SAMH… 0 0
40 7.86e-152 1.57e-148 Platelet -1.45 GZMM 0 0
41 1.63e-151 3.26e-148 Platelet -1.31 PPP2… 0 0
42 2.08e-151 4.17e-148 Platelet -1.17 APEX1 0 0
43 2.96e-151 5.92e-148 Platelet -1.22 XRCC5 0 0
44 4.10e-151 8.21e-148 Platelet -1.30 C9or… 0 0
45 6.44e-151 1.29e-147 Platelet -1.32 CD27 0 0
46 1.09e-149 2.18e-146 Platelet -1.27 SSBP1 0 0
47 1.33e-146 2.65e-143 Platelet -1.22 SEPT… 0 0
48 1.49e-144 2.97e-141 Platelet -1.16 MED10 0 0
49 5.10e-143 1.02e-139 Platelet -1.18 OCIA… 0 0
50 5.09e-142 1.02e-138 Platelet -1.17 TSTD1 0 0
51 7.34e-142 1.47e-138 Platelet -1.14 NOL7 0 0
52 2.09e-141 4.18e-138 Platelet -1.34 FAM2… 0 0
53 1.33e-140 2.67e-137 Platelet -1.29 CITE… 0 0
54 4.72e-140 9.44e-137 Platelet -1.18 RER1 0 0
55 1.11e-136 2.22e-133 Platelet -1.21 CD3G 0 0
56 3.28e-136 6.56e-133 Platelet -1.48 CEBPD 0 0
57 8.13e-135 1.63e-131 Platelet -1.09 RNF1… 0 0
58 1.71e-132 3.42e-129 Platelet -1.11 BUB3 0 0
59 3.65e-132 7.30e-129 Platelet -1.13 CLNS… 0 0
60 9.83e-132 1.97e-128 Platelet -1.26 APOB… 0 0
61 2.89e-131 5.78e-128 Platelet -1.06 TECR 0 0
62 9.68e-131 1.94e-127 Platelet -1.06 LSM10 0 0
63 2.24e-130 4.47e-127 Platelet -1.07 HSPE1 0 0
64 3.67e-130 7.34e-127 Platelet -1.07 CCT3 0 0
65 4.25e-129 8.50e-126 Platelet -1.06 CCT4 0 0
66 1.95e-128 3.91e-125 Platelet -1.17 FAIM3 0 0
67 8.66e-128 1.73e-124 Platelet -1.07 POLR… 0 0
68 9.39e-128 1.88e-124 Platelet -2.20 LGAL… 0 0
69 1.56e-127 3.12e-124 Platelet -1.06 GLIP… 0 0
70 1.95e-125 3.90e-122 Platelet -1.10 KARS 0 0
71 1.20e-124 2.40e-121 Platelet -1.06 NR4A2 0 0
72 4.53e-124 9.05e-121 Platelet -1.01 TRAP… 0 0
73 3.07e-123 6.15e-120 Platelet -1.02 SRRM2 0 0
74 3.45e-123 6.90e-120 Platelet -1.05 HSPA5 0 0
75 1.13e-120 2.26e-117 Platelet -1.03 EZR 0 0
76 1.20e-120 2.39e-117 Platelet -1.15 CCR7 0 0
77 1.23e-120 2.45e-117 Platelet -0.997 ENY2 0 0
78 5.43e-120 1.09e-116 Platelet -1.03 OCIA… 0 0
79 1.15e-119 2.30e-116 Platelet -1.06 EWSR1 0 0
80 7.50e-118 1.50e-114 Platelet -0.975 COPS6 0 0
81 1.28e-117 2.56e-114 Platelet -0.999 PRRC… 0 0
82 2.29e-117 4.58e-114 Platelet -0.982 DCK 0 0
83 3.76e-117 7.52e-114 Platelet -1.99 GZMA 0 0
84 3.56e-116 7.12e-113 Platelet -0.998 NSMC… 0 0
85 4.49e-115 8.97e-112 Platelet -2.02 CST7 0 0
86 5.37e-115 1.07e-111 Platelet -1.10 MYADM 0 0
87 3.44e-114 6.88e-111 Platelet -0.993 OSTC 0 0
88 7.21e-114 1.44e-110 Platelet -0.982 C1or… 0 0
89 7.51e-114 1.50e-110 Platelet -0.966 DECR1 0 0
90 8.64e-114 1.73e-110 Platelet -1.01 TAGAP 0 0
91 5.71e-113 1.14e-109 Platelet -0.958 MYLIP 0 0
92 1.02e-112 2.05e-109 Platelet -0.945 HMOX2 0 0
93 4.93e-112 9.86e-109 Platelet -0.962 PDCD4 0 0
94 8.90e-112 1.78e-108 Platelet -0.964 RPA2 0 0
95 7.08e-111 1.42e-107 Platelet -1.07 ZC3H… 0 0
96 7.88e-111 1.58e-107 Platelet -0.958 MRPS… 0 0
97 2.03e-109 4.07e-106 Platelet -0.966 C1or… 0 0
98 8.34e-109 1.67e-105 Platelet -1.18 HLA-… 0 0
99 1.08e-108 2.15e-105 Platelet -0.969 ANXA… 0 0
100 1.09e-108 2.18e-105 Platelet -1.04 IFI35 0 0
101 2.77e-108 5.54e-105 Platelet -1.05 HNRN… 0 0
102 3.21e-108 6.43e-105 Platelet -0.971 ARGL… 0 0
103 1.45e-107 2.90e-104 Platelet -0.985 CTSH 0 0
104 6.47e-107 1.29e-103 Platelet -0.986 TRADD 0 0
105 9.06e-107 1.81e-103 Platelet -0.930 HSPD1 0 0
106 3.37e-106 6.73e-103 Platelet -1.02 SOCS3 0 0
107 3.31e-105 6.63e-102 Platelet -0.977 GGNB… 0 0
108 4.86e-105 9.72e-102 Platelet -0.956 FLT3… 0 0
109 7.78e-105 1.56e-101 Platelet -0.929 IL16 0 0
110 5.36e-104 1.07e-100 Platelet -1.13 NCF2 0 0
111 6.33e-104 1.27e-100 Platelet -0.985 DDX17 0 0
112 1.05e-103 2.11e-100 Platelet -1.03 SURF1 0 0
113 3.21e-103 6.42e-100 Platelet -0.884 DGCR… 0 0
114 3.81e-103 7.61e-100 Platelet -0.961 PRKC… 0 0
115 7.45e-103 1.49e- 99 Platelet -0.980 CCDC… 0 0
116 9.49e-103 1.90e- 99 Platelet -0.908 NDUF… 0 0
117 1.34e-102 2.68e- 99 Platelet -0.920 MED28 0 0
118 2.13e-102 4.25e- 99 Platelet -0.929 LBH 0 0
119 1.01e-101 2.01e- 98 Platelet -0.891 RNF1… 0 0
120 1.53e-101 3.05e- 98 Platelet -0.889 FYN 0 0
121 8.82e-101 1.76e- 97 Platelet -0.916 RNAS… 0 0
122 9.72e-101 1.94e- 97 Platelet -0.922 FOXP1 0 0
123 9.75e-101 1.95e- 97 Platelet -0.969 RBM25 0 0
124 1.24e-100 2.49e- 97 Platelet -0.958 CWC15 0 0
125 2.25e-100 4.50e- 97 Platelet -0.883 SHKB… 0 0
126 5.13e-100 1.03e- 96 Platelet -0.881 EPC1 0 0
127 1.05e- 99 2.10e- 96 Platelet -0.860 TRAP… 0 0
128 1.80e- 99 3.60e- 96 Platelet -0.874 MRPS… 0 0
129 2.56e- 99 5.13e- 96 Platelet -1.08 NCF1 0 0
130 2.94e- 99 5.88e- 96 Platelet -0.876 SEC1… 0 0
131 9.08e- 99 1.82e- 95 Platelet -0.957 YPEL3 0 0
132 1.02e- 98 2.03e- 95 Platelet -0.887 PBXI… 0 0
133 3.43e- 98 6.86e- 95 Platelet -0.936 LSM6 0 0
134 3.61e- 98 7.21e- 95 Platelet -0.943 MRPS6 0 0
135 4.92e- 98 9.84e- 95 Platelet -0.871 ARHG… 0 0
136 7.01e- 98 1.40e- 94 Platelet -1.13 IFI30 0 0
137 8.72e- 98 1.74e- 94 Platelet -0.877 TNFA… 0 0
138 2.30e- 97 4.61e- 94 Platelet -0.927 DDX46 0 0
139 2.46e- 97 4.92e- 94 Platelet -0.890 DRAM2 0 0
140 5.18e- 97 1.04e- 93 Platelet -0.997 SH3K… 0 0
141 5.24e- 97 1.05e- 93 Platelet -0.926 OAZ2 0 0
142 7.96e- 97 1.59e- 93 Platelet -0.951 TRAP… 0 0
143 1.23e- 96 2.45e- 93 Platelet -0.961 SMIM7 0 0
144 3.00e- 96 5.99e- 93 Platelet -0.956 DENN… 0 0
145 1.10e- 95 2.19e- 92 Platelet -0.992 THOC7 0 0
146 1.97e- 95 3.95e- 92 Platelet -0.842 CMTM3 0 0
147 1.11e- 94 2.21e- 91 Platelet -1.02 PQBP1 0 0
148 5.59e- 94 1.12e- 90 Platelet -0.848 ZNF4… 0 0
149 1.40e- 93 2.80e- 90 Platelet -0.950 HDAC1 0 0
150 1.60e- 92 3.19e- 89 Platelet -0.901 ARL4A 0 0
151 1.92e- 92 3.84e- 89 Platelet -0.840 PIM2 0 0
152 4.51e- 92 9.02e- 89 Platelet -0.875 C17o… 0 0
153 6.21e- 92 1.24e- 88 Platelet -0.830 VIMP 0 0
154 1.40e- 91 2.80e- 88 Platelet -1.45 MS4A1 0 0
155 1.53e- 91 3.06e- 88 Platelet -1.38 MS4A… 0 0
156 2.89e- 91 5.78e- 88 Platelet -0.807 PSMA3 0 0
157 8.74e- 91 1.75e- 87 Platelet -0.872 TAF1D 0 0
158 2.02e- 90 4.04e- 87 Platelet -3.19 GNLY 0 0
159 6.80e- 90 1.36e- 86 Platelet -0.864 NME3 0 0
160 5.29e- 89 1.06e- 85 Platelet -0.879 TMEM… 0 0
161 2.26e- 88 4.51e- 85 Platelet -0.910 MYC 0 0
162 3.75e- 88 7.50e- 85 Platelet -0.821 NDUF… 0 0
163 4.25e- 88 8.50e- 85 Platelet -0.871 FAM1… 0 0
164 6.91e- 88 1.38e- 84 Platelet -0.812 SIT1 0 0
165 1.03e- 87 2.07e- 84 Platelet -0.817 ATM 0 0
166 7.67e- 87 1.53e- 83 Platelet -0.821 OPTN 0 0
167 8.02e- 87 1.60e- 83 Platelet -0.791 GPBP1 0 0
168 1.10e- 86 2.21e- 83 Platelet -0.785 NOP56 0 0
169 1.14e- 86 2.27e- 83 Platelet -1.24 HOPX 0 0
170 1.99e- 86 3.97e- 83 Platelet -0.817 POLR… 0 0
171 2.83e- 86 5.67e- 83 Platelet -0.819 BBX 0 0
172 3.07e- 86 6.14e- 83 Platelet -0.943 PCSK7 0 0
173 1.00e- 85 2.01e- 82 Platelet -0.788 METT… 0 0
174 1.56e- 85 3.11e- 82 Platelet -1.02 CDA 0 0
175 2.63e- 85 5.26e- 82 Platelet -0.880 NUP2… 0 0
176 4.19e- 85 8.38e- 82 Platelet -0.822 SF3B1 0 0
177 5.80e- 85 1.16e- 81 Platelet -0.807 DNAJ… 0 0
178 1.04e- 84 2.08e- 81 Platelet -1.22 CD14 0 0
179 2.22e- 84 4.43e- 81 Platelet -0.805 SARS 0 0
180 2.29e- 84 4.58e- 81 Platelet -0.842 ITGB7 0 0
181 2.49e- 84 4.98e- 81 Platelet -0.776 PLSC… 0 0
182 6.44e- 84 1.29e- 80 Platelet -0.788 SNW1 0 0
183 1.19e- 83 2.37e- 80 Platelet -0.777 MRPL… 0 0
184 1.25e- 83 2.49e- 80 Platelet -0.811 CAT 0 0
185 1.71e- 83 3.42e- 80 Platelet -0.807 PTGE… 0 0
186 2.40e- 83 4.80e- 80 Platelet -0.771 ARL2… 0 0
187 2.64e- 83 5.28e- 80 Platelet -0.926 ELOF1 0 0
188 1.83e- 82 3.66e- 79 Platelet -0.765 STAT1 0 0
189 3.80e- 82 7.59e- 79 Platelet -0.771 ZNF3… 0 0
190 1.33e- 81 2.65e- 78 Platelet -0.758 TMC8 0 0
191 9.95e- 81 1.99e- 77 Platelet -0.743 GLO1 0 0
192 9.95e- 81 1.99e- 77 Platelet -0.735 TTC3… 0 0
193 1.39e- 80 2.77e- 77 Platelet -0.735 PHF11 0 0
194 3.02e- 80 6.04e- 77 Platelet -0.805 NAA20 0 0
195 5.59e- 80 1.12e- 76 Platelet -0.762 RCN2 0 0
196 6.76e- 80 1.35e- 76 Platelet -0.739 SCO2 0 0
197 1.06e- 79 2.13e- 76 Platelet -0.878 MAL 0 0
198 1.19e- 79 2.39e- 76 Platelet -0.791 PRDX3 0 0
199 3.80e- 79 7.60e- 76 Platelet -0.823 CDC1… 0 0
200 6.38e- 79 1.28e- 75 Platelet -0.755 NKTR 0 0
201 6.60e- 79 1.32e- 75 Platelet -0.838 PMAI… 0 0
202 2.58e- 78 5.17e- 75 Platelet -0.743 LRCH4 0 0
203 2.87e- 78 5.75e- 75 Platelet -0.743 JAGN1 0 0
204 3.06e- 78 6.12e- 75 Platelet -1.59 CCL3 0 0
205 7.86e- 78 1.57e- 74 Platelet -0.833 BIRC3 0 0
206 1.37e- 77 2.73e- 74 Platelet -0.877 MMAD… 0 0
207 1.65e- 77 3.30e- 74 Platelet -0.744 KTN1 0 0
208 4.64e- 77 9.27e- 74 Platelet -0.807 RNF1… 0 0
209 5.34e- 77 1.07e- 73 Platelet -0.745 RP11… 0 0
210 2.67e- 76 5.34e- 73 Platelet -0.733 DDX6 0 0
211 3.77e- 76 7.54e- 73 Platelet -0.741 C20o… 0 0
212 9.26e- 76 1.85e- 72 Platelet -0.766 STOM… 0 0
213 1.05e- 75 2.10e- 72 Platelet -0.712 CD30… 0 0
214 1.52e- 75 3.04e- 72 Platelet -0.707 ATG12 0 0
215 1.73e- 75 3.47e- 72 Platelet -0.734 ANKR… 0 0
216 1.87e- 75 3.74e- 72 Platelet -0.699 DARS 0 0
217 8.35e- 75 1.67e- 71 Platelet -0.743 YTHD… 0 0
218 2.04e- 74 4.09e- 71 Platelet -0.748 SLC2… 0 0
219 2.10e- 74 4.20e- 71 Platelet -0.698 SLC3… 0 0
220 2.32e- 74 4.64e- 71 Platelet -0.719 EMC10 0 0
221 3.88e- 74 7.77e- 71 Platelet -0.711 ITPA 0 0
222 5.53e- 74 1.11e- 70 Platelet -0.707 NAPR… 0 0
223 5.63e- 74 1.13e- 70 Platelet -0.798 C5or… 0 0
224 1.49e- 73 2.99e- 70 Platelet -0.686 SORL1 0 0
225 3.16e- 73 6.32e- 70 Platelet -0.719 PPM1K 0 0
226 3.38e- 73 6.76e- 70 Platelet -0.769 TTC1 0 0
227 3.56e- 73 7.12e- 70 Platelet -0.695 ODF2L 0 0
228 4.54e- 73 9.08e- 70 Platelet -0.711 CCDC… 0 0
229 5.04e- 73 1.01e- 69 Platelet -0.680 EIF4… 0 0
230 8.76e- 73 1.75e- 69 Platelet -0.712 TTC3 0 0
231 1.11e- 72 2.22e- 69 Platelet -0.734 BABA… 0 0
232 1.46e- 72 2.92e- 69 Platelet -0.689 COMM… 0 0
233 2.78e- 72 5.55e- 69 Platelet -0.815 IRF9 0 0
234 5.56e- 72 1.11e- 68 Platelet -1.04 APOB… 0 0
235 5.77e- 72 1.15e- 68 Platelet -0.686 EIF4… 0 0
236 1.08e- 71 2.16e- 68 Platelet -0.693 CLPP 0 0
237 1.56e- 71 3.11e- 68 Platelet -0.907 HVCN1 0 0
238 2.19e- 71 4.37e- 68 Platelet -0.679 MRPL… 0 0
239 2.72e- 71 5.45e- 68 Platelet -0.809 GPR1… 0 0
240 3.84e- 71 7.69e- 68 Platelet -0.676 DHRS… 0 0
241 4.45e- 71 8.90e- 68 Platelet -0.718 KLF4 0 0
242 1.96e- 70 3.92e- 67 Platelet -0.767 COQ7 0 0
243 2.49e- 70 4.99e- 67 Platelet -0.681 CD96 0 0
244 2.69e- 70 5.38e- 67 Platelet -0.743 LILR… 0 0
245 3.92e- 70 7.84e- 67 Platelet -0.674 ADK 0 0
246 6.40e- 70 1.28e- 66 Platelet -0.691 NUDT5 0 0
247 1.18e- 69 2.37e- 66 Platelet -0.657 IMP4 0 0
248 1.27e- 69 2.54e- 66 Platelet -0.687 UBE2… 0 0
249 1.31e- 69 2.62e- 66 Platelet -0.906 CD8A 0 0
250 1.34e- 69 2.68e- 66 Platelet -0.705 RORA 0 0
251 2.00e- 69 4.00e- 66 Platelet -0.803 SAMD3 0 0
252 2.17e- 69 4.35e- 66 Platelet -0.686 SMIM… 0 0
253 2.46e- 69 4.93e- 66 Platelet -0.754 NEMF 0 0
254 2.94e- 69 5.89e- 66 Platelet -0.705 IDI1 0 0
255 3.70e- 69 7.40e- 66 Platelet -0.676 TMEM… 0 0
256 7.48e- 69 1.50e- 65 Platelet -0.707 ABT1 0 0
257 1.94e- 68 3.88e- 65 Platelet -0.651 ARID… 0 0
258 2.25e- 68 4.50e- 65 Platelet -0.700 BEX4 0 0
259 4.14e- 68 8.27e- 65 Platelet -0.670 IFT20 0 0
260 5.39e- 68 1.08e- 64 Platelet -0.695 SH3B… 0 0
261 6.11e- 68 1.22e- 64 Platelet -0.673 PSTP… 0 0
262 6.67e- 68 1.33e- 64 Platelet -0.731 ARL2 0 0
263 7.93e- 68 1.59e- 64 Platelet -0.775 CSF3R 0 0
264 8.03e- 68 1.61e- 64 Platelet -0.795 PARP1 0 0
265 9.90e- 68 1.98e- 64 Platelet -0.662 MRPS… 0 0
266 2.14e- 67 4.28e- 64 Platelet -0.646 THRA… 0 0
267 4.24e- 67 8.48e- 64 Platelet -0.891 KLRG1 0 0
268 9.86e- 67 1.97e- 63 Platelet -0.666 MRPS… 0 0
269 1.03e- 66 2.07e- 63 Platelet -0.678 CIR1 0 0
270 1.21e- 66 2.42e- 63 Platelet -0.636 CYB5… 0 0
271 8.57e- 66 1.71e- 62 Platelet -0.666 DNAJ… 0 0
272 8.89e- 66 1.78e- 62 Platelet -0.738 TMEM… 0 0
273 1.54e- 65 3.07e- 62 Platelet -0.836 HAGH 0 0
274 1.74e- 65 3.49e- 62 Platelet -0.637 ARHG… 0 0
275 3.08e- 65 6.17e- 62 Platelet -0.650 IFI44 0 0
276 3.63e- 65 7.25e- 62 Platelet -0.633 TXND… 0 0
277 6.37e- 65 1.27e- 61 Platelet -0.643 BEX2 0 0
278 1.27e- 64 2.54e- 61 Platelet -0.675 AATF 0 0
279 1.30e- 64 2.60e- 61 Platelet -0.670 GIT2 0 0
280 2.12e- 64 4.23e- 61 Platelet -0.794 IRF8 0 0
281 4.16e- 64 8.31e- 61 Platelet -0.825 ALDH2 0 0
282 5.44e- 64 1.09e- 60 Platelet -0.624 STMN3 0 0
283 6.22e- 64 1.24e- 60 Platelet -0.665 TMEM… 0 0
284 8.53e- 64 1.71e- 60 Platelet -0.701 RPL7… 0 0
285 1.03e- 63 2.06e- 60 Platelet -0.635 RGS14 0 0
286 1.41e- 63 2.82e- 60 Platelet -0.728 ARID… 0 0
287 2.31e- 63 4.62e- 60 Platelet -0.613 USE1 0 0
288 3.65e- 63 7.31e- 60 Platelet -0.740 ECHD… 0 0
289 4.37e- 63 8.75e- 60 Platelet -0.697 NUDT… 0 0
290 4.63e- 63 9.26e- 60 Platelet -0.634 SH3B… 0 0
291 5.07e- 63 1.01e- 59 Platelet -0.698 NR3C1 0 0
292 5.50e- 63 1.10e- 59 Platelet -0.633 CD40… 0 0
293 6.27e- 63 1.25e- 59 Platelet -0.654 FKBP3 0 0
294 7.96e- 63 1.59e- 59 Platelet -0.628 IL27… 0 0
295 1.93e- 62 3.87e- 59 Platelet -0.708 IGSF6 0 0
296 1.96e- 62 3.93e- 59 Platelet -1.49 CCL4 0 0
297 3.42e- 62 6.85e- 59 Platelet -0.703 C21o… 0 0
298 5.47e- 62 1.09e- 58 Platelet -0.631 PRNP 0 0
299 5.80e- 62 1.16e- 58 Platelet -0.641 SNX5 0 0
300 7.54e- 62 1.51e- 58 Platelet -0.608 FRG1B 0 0
# … with 468 more rows, and 1 more variable: count <dbl>volcano_plot <- mgs_t_platelet %>%
mutate(log_p = -log(p_value, base = 10)) %>%
mutate(is_zero = count == 0) %>%
ggplot(aes(log_fc, log_p, colour = factor(is_zero))) +
scale_colour_manual(values = c("black", "red")) +
geom_point() +
labs(
y = "-log10(p-value)",
x = "Log fold-change",
colour = "All zero expression",
title = "Platelet cluster, pbmc3k data"
) +
theme_bw()
volcano_plot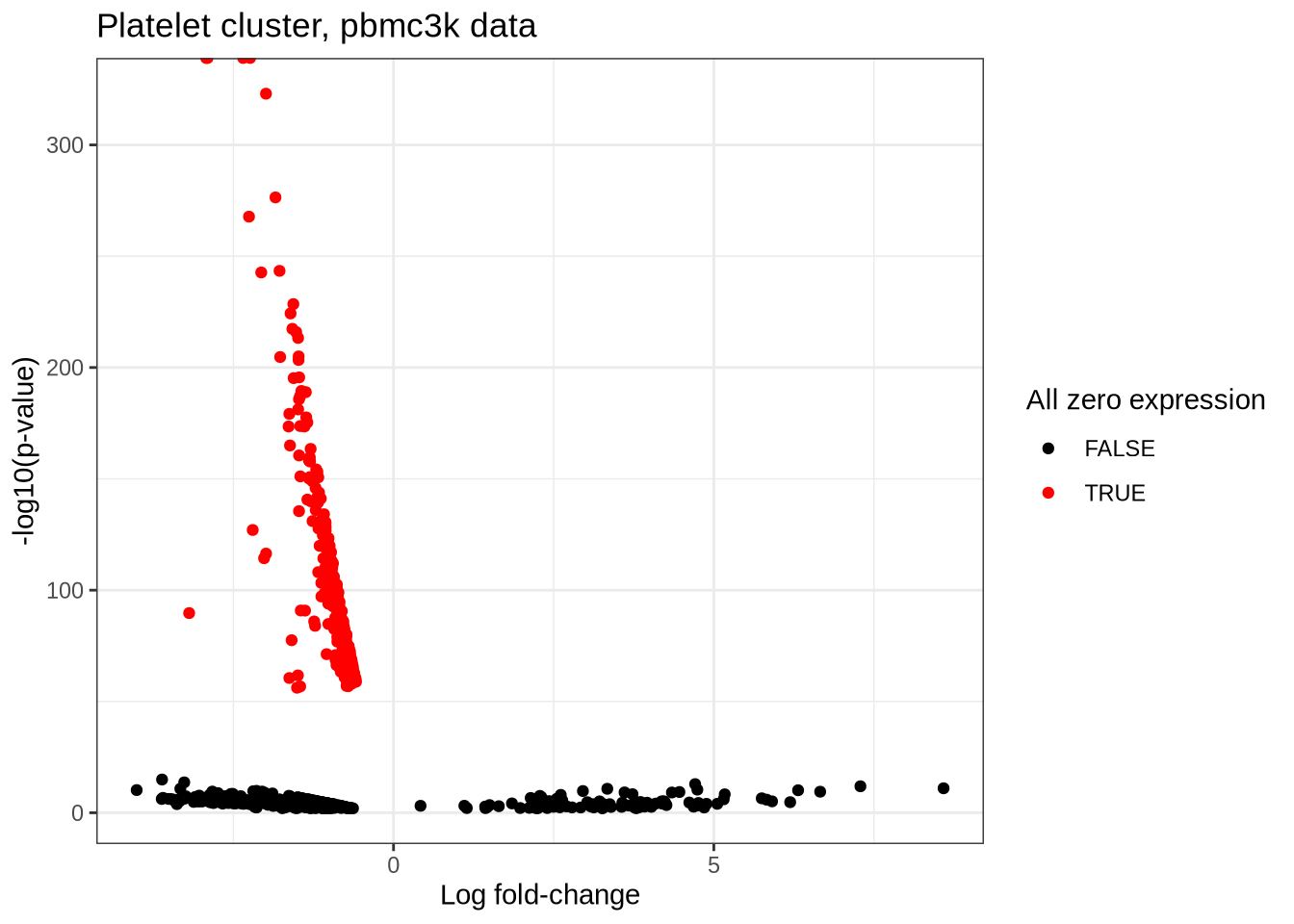
saveRDS(
volcano_plot,
here::here("figures", "raw", "volcano-plot.rds"),
)rank_log_fc_plot <- mgs_t_platelet %>%
mutate(is_zero = count == 0) %>%
mutate(rank = 1:n()) %>%
ggplot(aes(x = rank, y = log_fc, colour = factor(is_zero))) +
geom_point() +
geom_vline(xintercept = 284) +
coord_cartesian(ylim = c(-5, NA)) +
labs(
x = "Rank of gene",
y = "Log fold-change value",
colour = "All zeros",
title = "Rank vs log fold change"
) +
theme_bw()
rank_log_fc_plot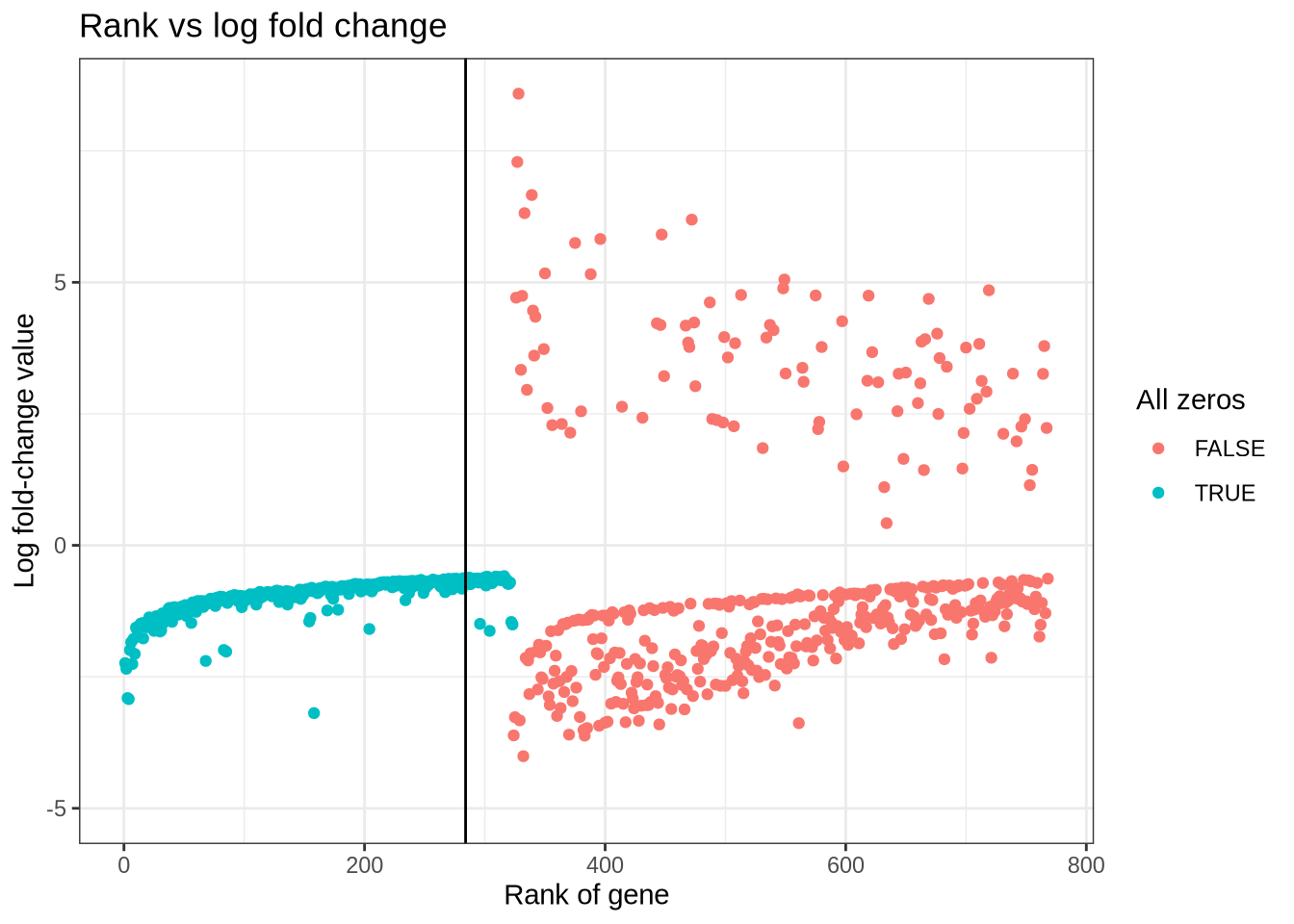
saveRDS(
rank_log_fc_plot,
here::here("figures", "raw", "rank-log-fc.rds")
)wrap_plots(rank_log_fc_plot, volcano_plot, guides = "collect") +
plot_annotation(tag_levels = "a") &
theme(plot.tag = element_text(size = 18))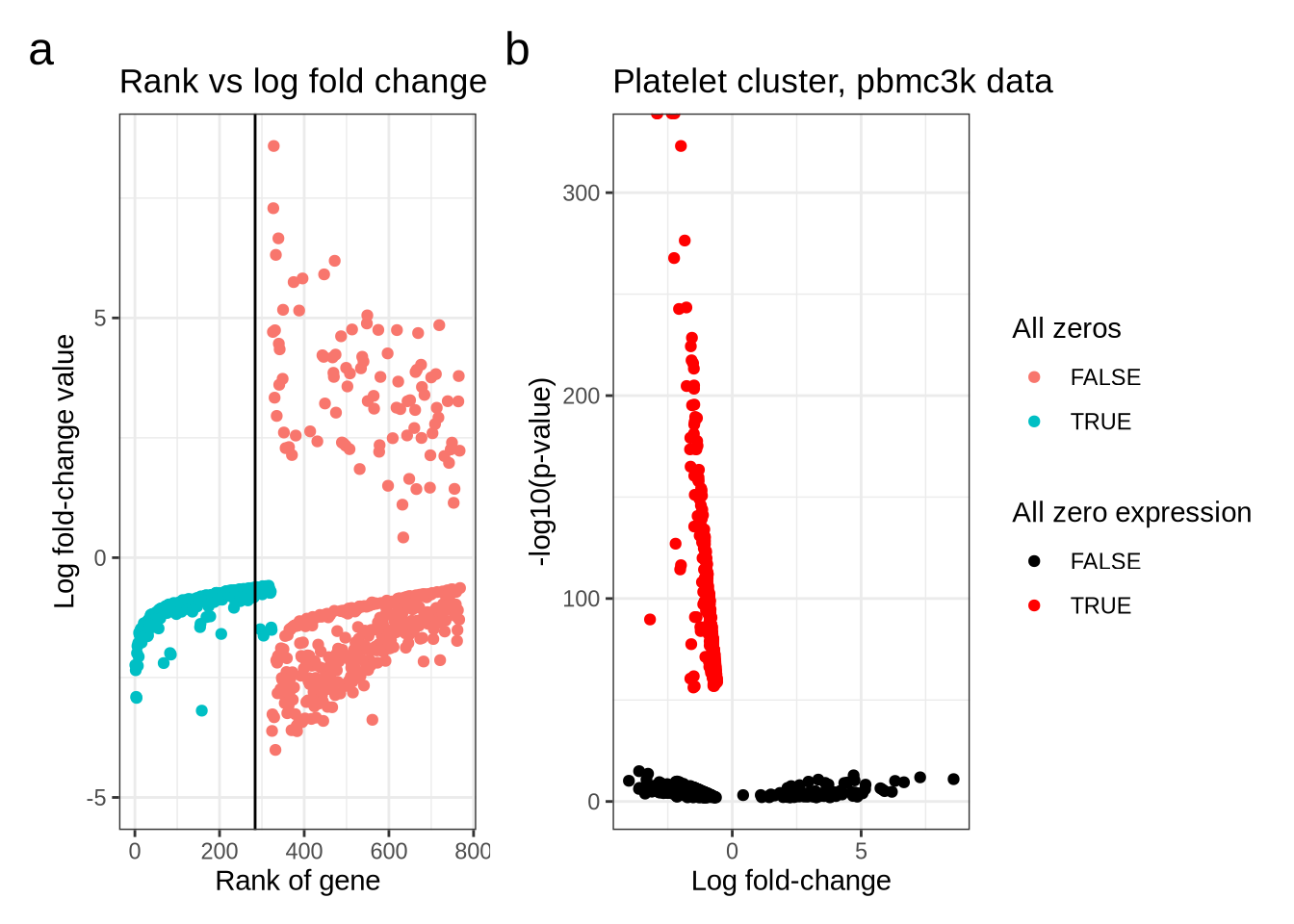
cell_types <- unique(pbmc3k$label)
zero_gene_counts <- lapply(cell_types, function(x) {
counts <- rowSums(counts(pbmc3k)[, colLabels(pbmc3k) == x])
sum(counts == 0)
})
names(zero_gene_counts) <- unique(pbmc3k$label)
zero_gene_counts$`Memory CD4 T`
[1] 17
$B
[1] 23
$`CD14+ Mono`
[1] 8
$NK
[1] 56
$`CD8 T`
[1] 29
$`FCGR3A+ Mono`
[1] 25
$`Naive CD4 T`
[1] 12
$DC
[1] 83
$Platelet
[1] 720endo_cell_types <- unique(endothelial$label)
lapply(endo_cell_types, function(x) {
counts <- rowSums(counts(endothelial)[, colLabels(endothelial) == x])
sum(counts == 0)
})[[1]]
[1] 0
[[2]]
[1] 0
[[3]]
[1] 0
[[4]]
[1] 1
[[5]]
[1] 2
[[6]]
[1] 0
[[7]]
[1] 24lawlor_cell_types <- unique(lawlor$label)
lawlor_zero_gene_counts <- lapply(lawlor_cell_types, function(x) {
counts <- rowSums(counts(lawlor)[, colLabels(lawlor) == x])
sum(counts == 0)
})
names(lawlor_zero_gene_counts) <- unique(lawlor$label)
lawlor_zero_gene_counts$Beta
[1] 0
$Stellate
[1] 50
$Ductal
[1] 17
$Alpha
[1] 0
$Acinar
[1] 24
$`None/Other`
[1] 14
$`Gamma/PP`
[1] 11
$Delta
[1] 4mgs_t_raw %>%
pluck("result") %>%
filter(cluster == "NK")# A tibble: 663 × 7
p_value p_value_adj cluster log_fc gene raw_statistic scaled_statistic
<dbl> <dbl> <fct> <dbl> <chr> <dbl> <dbl>
1 2.57e-240 5.13e-237 NK 4.04 NKG7 0 0
2 2.28e- 98 4.55e- 95 NK -1.17 IFI30 0 0
3 2.70e- 98 5.41e- 95 NK 2.91 CTSW 0 0
4 1.95e- 91 3.90e- 88 NK 1.22 HLA-C 0 0
5 7.64e- 89 1.53e- 85 NK 0.937 B2M 0 0
6 1.25e- 87 2.51e- 84 NK 5.32 GNLY 0 0
7 9.78e- 87 1.96e- 83 NK 4.83 GZMB 0 0
8 1.69e- 83 3.37e- 80 NK -1.20 LY86 0 0
9 1.13e- 81 2.26e- 78 NK -0.890 SLC7A7 0 0
10 3.03e- 78 6.06e- 75 NK 4.00 PRF1 0 0
# … with 653 more rowswhich(rowSums(counts(pbmc3k)[, colLabels(pbmc3k) == "NK"]) == 0) PPBP PF4 GNG11 IFI30 VPREB3
183 257 302 306 337
CLU TUBB1 CD9 RBP7 RP11-290F20.3
365 402 404 442 459
FPR1 SLC7A7 SPARC APOBEC3B FOLR3
481 515 528 530 544
GP9 PLBD1 CA2 PTCRA BLK
557 558 672 714 760
ACRBP ITGA2B RETN ARHGAP24 PPP1R14A
821 871 895 904 933
AL928768.3 SPIB RAB32 SNN BLNK
1038 1057 1118 1121 1177
GAPT C16orf74 YPEL2 TMEM40 C19orf38
1179 1209 1212 1222 1306
KCNG1 BACE2 GBGT1 CD19 LILRA3
1308 1355 1371 1404 1428
RP11-291B21.2 CD33 SCAPER CMTM5 FCGR1A
1430 1506 1581 1614 1639
CDKN2A F13A1 C1QA SCGB3A1 GMPR
1716 1732 1740 1765 1768
OSCAR CLDN5 MTRF1 PHACTR1 LRRIQ3
1790 1792 1825 1877 1925
AP001189.4
1969 n_1 <- 1000
n_2 <- 40
y_1 <- rnorm(n_1, 3, 2)
y_2 <- rnorm(n_2, 2, 1)
calculate_nu <- function(y_1, y_2) {
s2_1 <- stats::var(y_1)
s2_2 <- stats::var(y_2)
n_1 <- length(y_1)
n_2 <- length(y_2)
num <- (s2_1 / n_1 + s2_2 / n_2)^2
denom <- (s2_1 / n_1)^2 / (n_1 - 1) + (s2_2 / n_2)^2 / (n_2 - 1)
num / denom
}
nu <- numeric(n_2)
for (i in seq_len(n_2)) {
y_2[1:i] <- 0
nu[[i]] <- calculate_nu(y_1, y_2)
}
tibble(n_zero = 1:n_2, nu) %>%
ggplot(aes(x = n_zero, y = nu)) +
geom_point() +
labs(
x = "Number of zeros",
y = "Degrees of freedom"
) +
theme_bw()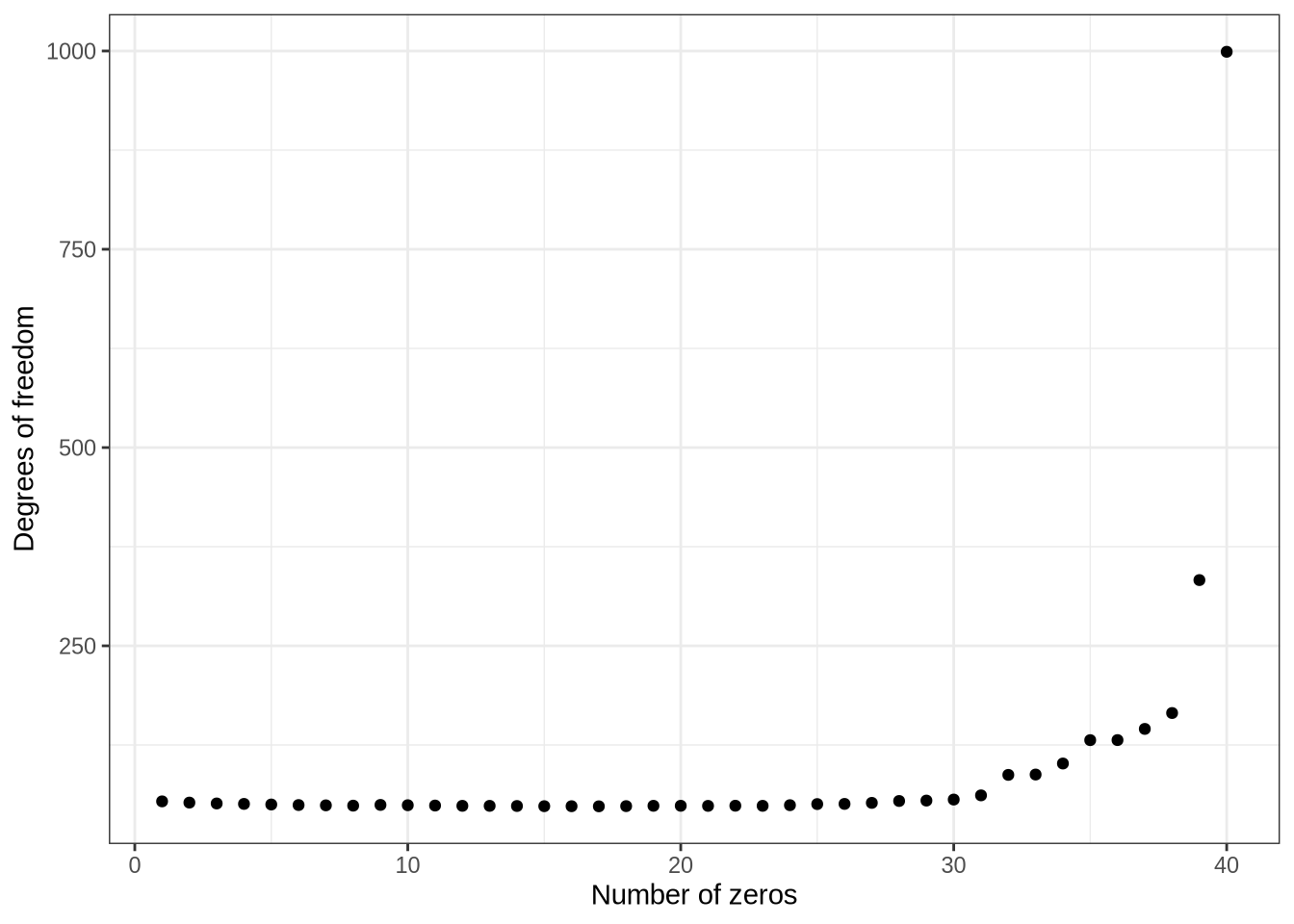
X_bar <- 10
S2 <- 10
X <- as.vector(logcounts(pbmc3k["LCK", pbmc3k$label != "Platelet"]))
X_bar <- mean(X)
S2_X <- stats::var(X)
n_X <- length(X)
T <- X_bar / sqrt(S2_X / n_X)
nu <- n_X - 1
2 * pt(T, df = nu, lower.tail = FALSE)[1] 3.744449e-244Y <- c(1, rep(0, 13))
Y_bar <- mean(Y)
S2_Y <- stats::var(Y)
n_Y <- length(Y)
T_prime <- (X_bar - Y_bar) / sqrt(S2_X / n_X + S2_Y / n_Y)
2 * pt(T_prime, df = nu, lower.tail = FALSE)[1] 1.811676e-17nu_prime <- calculate_nu(X, Y)
2 * pt(T_prime, df = nu_prime, lower.tail = FALSE)[1] 3.895889e-07
devtools::session_info()─ Session info ──────────────────────────────────────────────────────────────
hash: flag: Zambia, hatching chick, flag: Gabon
setting value
version R version 4.1.2 (2021-11-01)
os Red Hat Enterprise Linux 9.2 (Plow)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2024-01-01
pandoc 2.18 @ /apps/easybuild-2022/easybuild/software/MPI/GCC/11.3.0/OpenMPI/4.1.4/RStudio-Server/2022.07.2+576-Java-11-R-4.1.2/bin/pandoc/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
assertthat 0.2.1 2019-03-21 [2] CRAN (R 4.1.2)
Biobase * 2.54.0 2021-10-26 [1] Bioconductor
BiocGenerics * 0.40.0 2021-10-26 [1] Bioconductor
bitops 1.0-7 2021-04-24 [2] CRAN (R 4.1.2)
bslib 0.3.1 2021-10-06 [1] CRAN (R 4.1.0)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.1.0)
callr 3.7.0 2021-04-20 [2] CRAN (R 4.1.2)
cli 3.6.1 2023-03-23 [1] CRAN (R 4.1.0)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.1.0)
crayon 1.5.1 2022-03-26 [1] CRAN (R 4.1.0)
DBI 1.1.2 2021-12-20 [1] CRAN (R 4.1.0)
DelayedArray 0.20.0 2021-10-26 [1] Bioconductor
desc 1.4.0 2021-09-28 [2] CRAN (R 4.1.2)
devtools 2.4.2 2021-06-07 [2] CRAN (R 4.1.2)
digest 0.6.29 2021-12-01 [1] CRAN (R 4.1.0)
dplyr * 1.0.9 2022-04-28 [1] CRAN (R 4.1.0)
ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.2)
evaluate 0.14 2019-05-28 [2] CRAN (R 4.1.2)
fansi 1.0.4 2023-01-22 [1] CRAN (R 4.1.0)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.1.0)
fastmap 1.1.0 2021-01-25 [2] CRAN (R 4.1.2)
fs 1.5.2 2021-12-08 [1] CRAN (R 4.1.0)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.1.0)
GenomeInfoDb * 1.30.0 2021-10-26 [1] Bioconductor
GenomeInfoDbData 1.2.7 2021-12-03 [1] Bioconductor
GenomicRanges * 1.46.1 2021-11-18 [1] Bioconductor
ggplot2 * 3.3.6 2022-05-03 [1] CRAN (R 4.1.0)
git2r 0.28.0 2021-01-10 [2] CRAN (R 4.1.2)
glue 1.6.0 2021-12-17 [1] CRAN (R 4.1.0)
gtable 0.3.0 2019-03-25 [2] CRAN (R 4.1.2)
here 1.0.1 2020-12-13 [1] CRAN (R 4.1.0)
highr 0.9 2021-04-16 [2] CRAN (R 4.1.2)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
httpuv 1.6.5 2022-01-05 [1] CRAN (R 4.1.0)
IRanges * 2.28.0 2021-10-26 [1] Bioconductor
jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.2)
jsonlite 1.8.0 2022-02-22 [1] CRAN (R 4.1.0)
knitr 1.36 2021-09-29 [1] CRAN (R 4.1.0)
labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.2)
later 1.3.0 2021-08-18 [1] CRAN (R 4.1.0)
lattice 0.20-45 2021-09-22 [2] CRAN (R 4.1.2)
lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.1.0)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.1.0)
Matrix 1.3-4 2021-06-01 [2] CRAN (R 4.1.2)
MatrixGenerics * 1.6.0 2021-10-26 [1] Bioconductor
matrixStats * 0.62.0 2022-04-19 [1] CRAN (R 4.1.0)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.1.0)
munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.2)
patchwork * 1.1.1 2020-12-17 [2] CRAN (R 4.1.2)
pillar 1.7.0 2022-02-01 [1] CRAN (R 4.1.0)
pkgbuild 1.2.0 2020-12-15 [2] CRAN (R 4.1.2)
pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.2)
pkgload 1.2.3 2021-10-13 [2] CRAN (R 4.1.2)
prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.2)
processx 3.5.2 2021-04-30 [2] CRAN (R 4.1.2)
promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.2)
ps 1.7.1 2022-06-18 [1] CRAN (R 4.1.0)
purrr * 0.3.4 2020-04-17 [2] CRAN (R 4.1.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
Rcpp 1.0.8.3 2022-03-17 [1] CRAN (R 4.1.0)
RCurl 1.98-1.5 2021-09-17 [1] CRAN (R 4.1.0)
remotes 2.4.2 2021-11-30 [1] CRAN (R 4.1.0)
rlang 1.0.3 2022-06-27 [1] CRAN (R 4.1.0)
rmarkdown 2.14 2022-04-25 [1] CRAN (R 4.1.0)
rprojroot 2.0.3 2022-04-02 [1] CRAN (R 4.1.0)
rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.1.0)
S4Vectors * 0.32.3 2021-11-21 [1] Bioconductor
sass 0.4.1 2022-03-23 [1] CRAN (R 4.1.0)
scales 1.2.1 2022-08-20 [1] CRAN (R 4.1.0)
sessioninfo 1.2.0 2021-10-31 [2] CRAN (R 4.1.2)
SingleCellExperiment * 1.16.0 2021-10-26 [1] Bioconductor
stringi 1.7.6 2021-11-29 [1] CRAN (R 4.1.0)
stringr 1.4.0 2019-02-10 [2] CRAN (R 4.1.2)
SummarizedExperiment * 1.24.0 2021-10-26 [1] Bioconductor
testthat 3.1.0 2021-10-04 [2] CRAN (R 4.1.2)
tibble * 3.1.7 2022-05-03 [1] CRAN (R 4.1.0)
tidyselect 1.1.2 2022-02-21 [1] CRAN (R 4.1.0)
usethis 2.1.3 2021-10-27 [2] CRAN (R 4.1.2)
utf8 1.2.3 2023-01-31 [1] CRAN (R 4.1.0)
vctrs 0.4.1 2022-04-13 [1] CRAN (R 4.1.0)
whisker 0.4 2019-08-28 [2] CRAN (R 4.1.2)
withr 2.5.0 2022-03-03 [1] CRAN (R 4.1.0)
workflowr 1.7.0 2021-12-21 [1] CRAN (R 4.1.0)
xfun 0.31 2022-05-10 [1] CRAN (R 4.1.0)
XVector 0.34.0 2021-10-26 [1] Bioconductor
yaml 2.3.5 2022-02-21 [1] CRAN (R 4.1.0)
zlibbioc 1.40.0 2021-10-26 [1] Bioconductor
[1] /home/jpullin/R/x86_64-pc-linux-gnu-library/4.1
[2] /apps/easybuild-2022/easybuild/software/MPI/GCC/11.3.0/OpenMPI/4.1.4/R/4.1.2/lib64/R/library
──────────────────────────────────────────────────────────────────────────────