10x-Neuro: manual splatPop simulation
Christina B. Azodi
2021-11-19
Last updated: 2021-11-19
Checks: 6 1
Knit directory: KEJP_2020_splatPop/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210215) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0d55650. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .cache/
Ignored: .config/
Ignored: .snakemake/
Ignored: data/.DS_Store
Ignored: data/10x_lung/
Ignored: data/ALL.chr2.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.EURO.0.99.MAF05.filtered.vcf
Ignored: data/D30.h5
Ignored: data/Diabetes/
Ignored: data/IBD/
Ignored: data/agg_10X-fibroblasts-control.rds
Ignored: data/agg_IBD-ss2.rds
Ignored: data/agg_Neuro-10x_DA.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6.rds
Ignored: data/agg_diabetes-ss2.rds
Ignored: data/agg_iPSC-ss2_D0.rds
Ignored: data/covid/
Ignored: data/cuomo_NeuroSeq_10x_all_sce.rds
Ignored: data/iPSC_eqtl-mapping/
Ignored: data/lung_fibrosis/
Ignored: data/pseudoB_IBD-ss2.rds
Ignored: data/pseudoB_Neuro-10x_DA.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6.rds
Ignored: data/pseudoB_diabetes-ss2.rds
Ignored: data/pseudoB_iPSC-ss2_D0.rds
Ignored: data/sce_10X-fibroblasts-947170.rds
Ignored: data/sce_10X-fibroblasts-allGenes.rds
Ignored: data/sce_10X-fibroblasts.rds
Ignored: data/sce_IBD-ss2.rds
Ignored: data/sce_IBD-ss2_HC2.rds
Ignored: data/sce_Neuro-10x.rds
Ignored: data/sce_Neuro-10x_2CT.rds
Ignored: data/sce_Neuro-10x_DA-wihj4.rds
Ignored: data/sce_Neuro-10x_DA_allGenes.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6.rds
Ignored: data/sce_diabetes-ss2.rds
Ignored: data/sce_diabetes-ss2_T2D-5.rds
Ignored: data/sce_iPSC-ss2_D0-allGenes.rds
Ignored: data/sce_iPSC-ss2_D0-joxm39.rds
Ignored: data/sce_iPSC-ss2_D0.rds
Ignored: docs/.DS_Store
Ignored: output/00_Figures/
Ignored: output/01_sims/
Ignored: output/10x_eQTL-mapping/
Ignored: output/demo_eQTL/
Ignored: output/iPSC_eQTL/
Ignored: references/1000GP_Phase3_sample_info.txt
Ignored: references/Homo_sapiens.GRCh38.99.chromosome.22.gff3
Ignored: references/chr2.filt.2-temporary.bed
Ignored: references/chr2.filt.2-temporary.bim
Ignored: references/chr2.filt.2-temporary.fam
Ignored: references/chr2.filt.2.log
Ignored: references/chr2.filt.log
Ignored: references/chr2.filt.map
Ignored: references/chr2.filt.nosex
Ignored: references/chr2.filt.ped
Ignored: references/chr2.filt.prune.in
Ignored: references/chr2.filt.prune.out
Ignored: references/chr2.filtered.log
Ignored: references/chr2.filtered.nosex
Ignored: references/chr2.filtered.vcf
Ignored: references/chr2.filtered2.log
Ignored: references/chr2.filtered2.nosex
Ignored: references/chr2.filtered2.vcf
Ignored: references/chr2.filtered3.log
Ignored: references/chr2.filtered3.nosex
Ignored: references/chr2.filtered3.vcf
Ignored: references/chr2.genes.gff3
Ignored: references/chr2.vcf.gz
Ignored: references/chr22.filt.log
Ignored: references/chr22.filt.map
Ignored: references/chr22.filt.nosex
Ignored: references/chr22.filt.ped
Ignored: references/chr22.filt.prune.in
Ignored: references/chr22.filt.prune.out
Ignored: references/chr22.filtered.log
Ignored: references/chr22.filtered.nosex
Ignored: references/chr22.filtered.vcf
Ignored: references/chr22.filtered.vcf.bed
Ignored: references/chr22.filtered.vcf.bim
Ignored: references/chr22.filtered.vcf.fam
Ignored: references/chr22.filtered.vcf.log
Ignored: references/chr22.filtered.vcf.nosex
Ignored: references/chr22.filtered.vcf.rel
Ignored: references/chr22.filtered.vcf.rel.id
Ignored: references/chr22.filtered.vcf.rel_mod
Ignored: references/chr22.genes.gff3
Ignored: references/chr22.genes.gff3_annotation
Ignored: references/chr22.genes.gff3_chunks
Ignored: references/chr22.vcf.gz
Ignored: references/eqtl_summary_stats.tar.gz
Ignored: references/eqtl_summary_stats_renamed/
Ignored: references/keep_samples.txt
Ignored: references/remove_snps.txt
Ignored: references/test.genome
Ignored: references/test.log
Ignored: references/test.nosex
Untracked files:
Untracked: KEJP_2020_splatPop.Rproj
Unstaged changes:
Modified: .gitignore
Modified: .gitlab-ci.yml
Modified: CITATION
Modified: Dockerfile
Modified: LICENSE
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/10x-Neuro_emp-simulations.Rmd
Modified: analysis/10x-Neuro_estimate-params.Rmd
Modified: analysis/KEJP_iPSC-ss2.Rmd
Modified: analysis/_site.yml
Modified: analysis/about.Rmd
Modified: analysis/index.Rmd
Modified: analysis/license.Rmd
Modified: analysis/ss2-iPSC_estimate-params.Rmd
Modified: analysis/ss2-iPSC_simulations.Rmd
Modified: cluster.json
Modified: code/1_process-empirical-data.R
Modified: code/2_estimate-splatPopParams.R
Modified: code/4_simulate_DEG.R
Modified: code/README.md
Modified: code/plot_functions.R
Modified: code/plot_functions2.R
Modified: data/README.md
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.css
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.js
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cerulean.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cosmo.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/darkly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/flatly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Lato.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycle.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycleBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSans.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBoldItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLightItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Raleway.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RalewayBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Roboto.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoMedium.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansPro.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Ubuntu.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/journal.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/lumen.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/paper.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/readable.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/sandstone.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/simplex.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/spacelab.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/united.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/yeti.min.css
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.eot
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.svg
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.ttf
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff2
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.js
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.min.js
Modified: docs/site_libs/bootstrap-3.3.5/js/npm.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/html5shiv.min.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/respond.min.js
Modified: docs/site_libs/font-awesome-5.1.0/css/all.css
Modified: docs/site_libs/font-awesome-5.1.0/css/v4-shims.css
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff2
Modified: docs/site_libs/header-attrs-2.5/header-attrs.js
Modified: docs/site_libs/jquery-1.11.3/jquery.min.js
Modified: docs/site_libs/jqueryui-1.11.4/README
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_444444_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_555555_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777620_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777777_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_cc0000_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_ffffff_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/index.html
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.min.css
Modified: docs/site_libs/navigation-1.1/codefolding.js
Modified: docs/site_libs/navigation-1.1/sourceembed.js
Modified: docs/site_libs/navigation-1.1/tabsets.js
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.css
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.js
Modified: environment.yml
Modified: envs/limix_env.yaml
Modified: envs/myenv.yaml
Modified: org/README.md
Modified: org/project_management.org
Modified: output/README.md
Modified: references/README.md
Modified: resources/README.md
Modified: resources/keep_samples.txt
Modified: workflow/config_bp_ss2.yaml
Modified: workflow/sims_limix_v1.15.1.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/10x-Neuro_emp-simulations.Rmd) and HTML (public/10x-Neuro_emp-simulations.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0d55650 | cazodi | 2021-11-19 | update figure 4 to have scatter plots |
| html | 0d55650 | cazodi | 2021-11-19 | update figure 4 to have scatter plots |
| Rmd | 116e4a3 | cazodi | 2021-10-08 | update mean-variance plots for manuscript |
| Rmd | 7f8fd63 | cazodi | 2021-09-27 | add empirical replication example |
| html | 7f8fd63 | cazodi | 2021-09-27 | add empirical replication example |
| Rmd | f2bf14a | cazodi | 2021-09-21 | update eqtl and de results |
#install.packages("code/splatter_1.17.2.tar.gz", repos = NULL, type="source")
suppressPackageStartupMessages({
library(SingleCellExperiment)
library(scater)
library(tidyverse)
#detach("package:splatter", unload=TRUE)
library(splatter)
library(VariantAnnotation)
library(cluster)
library(fitdistrplus)
library(RColorBrewer)
library(ComplexHeatmap)
library(ggpubr)
library(data.table)
})
source("code/plot_functions.R")
source("code/misc_functions.R")date <- Sys.Date()
set.seed(42)
save <- FALSE
rerun <- FALSE
date.use <- "2021-11-18"
sample.colors <- projectColors("samples")
col_fun <- circlize::colorRamp2(c(0.85, 1), c("white", "red"))Using splatPop to replicate empirical data
The splatPop simulation approach involves estimating key parameters from real data and using those parameters to randomly sample values needed to simulate single-cell RNA-seq data for the desired number of genes, individuals, and cells. This results in a simulated dataset with similar properties as the empirical data, but not a replication of the empirical dataset. For example, the genes simulated by splatPop are not real genes and thus, gene-gene relationships are not maintained.
However, because of the flexible design of splatPop, there are a number of ways users can more closely replicate their read data if desired. Here we will show examples of using splatPop to:
- Replicate real eQTL and population level gene mean expression properties by providing a splatPop key.
- Replicate real eQTL and individual level gene mean expression values by providing empirical data.
We will base our simulations on the 10x-Neuro data, described here.
Input data
To run the default splatPop simulation pipeline, only genotype (vcf) and gene (gff) data and splatPop parameters need to be provided:
Genotype data
vcf <- readVcf("data/iPSC_eqtl-mapping/hipsci.chr22.vcf", "hg38")
vcf <- vcf[-which(duplicated(rownames(vcf)))]
geno(vcf)$GT <- gsub("/", "|", geno(vcf)$GT, fixed=TRUE)
colnames(vcf) <- gsub(".*-", "", colnames(vcf))
colnames(vcf) <- gsub("_.*", "", colnames(vcf))
geno(vcf)$GT[1:5,1:5] xugn wigw deyz lexy vabj
rs145711952 "0|0" "0|0" "0|0" "0|0" "0|0"
rs5746647 "1|1" "1|1" "1|1" "1|1" "1|1"
rs5747999 "0|1" "1|1" "1|1" "1|0" "1|1"
rs4428101 "0|0" "0|0" "0|0" "0|0" "0|0"
rs13056782 "0|0" "0|0" "0|0" "0|0" "0|0"Gene information
gff <- read.table("references/chr22.genes.gff3", sep="\t", header=FALSE, quote="")
geneKey <- as.data.frame(list(symbol=gsub(";.*", "", gsub(".*Name=", "", gff$V9)),
feature_id=gsub(".*:", "", gsub(";.*", "", gff$V9))))
geneKey <- geneKey[!duplicated(geneKey$symbol), ]
row.names(geneKey) <- geneKey$symbol
head(gff) V1 V2 V3 V4 V5 V6 V7 V8
1 22 ensembl_havana gene 15528159 15529139 . + .
2 22 ensembl_havana gene 15690026 15721631 . + .
3 22 ensembl_havana gene 16590751 16592810 . - .
4 22 ensembl_havana gene 16783412 16821699 . - .
5 22 ensembl_havana gene 16961936 17008222 . - .
6 22 ensembl_havana gene 17084954 17115694 . + .
V9
1 ID=gene:ENSG00000130538;Name=OR11H1;biotype=protein_coding;description=olfactory receptor family 11 subfamily H member 1 [Source:HGNC Symbol%3BAcc:HGNC:15404];gene_id=ENSG00000130538;logic_name=ensembl_havana_gene_homo_sapiens;version=5
2 ID=gene:ENSG00000198062;Name=POTEH;biotype=protein_coding;description=POTE ankyrin domain family member H [Source:HGNC Symbol%3BAcc:HGNC:133];gene_id=ENSG00000198062;logic_name=ensembl_havana_gene_homo_sapiens;version=15
3 ID=gene:ENSG00000198445;Name=CCT8L2;biotype=protein_coding;description=chaperonin containing TCP1 subunit 8 like 2 [Source:HGNC Symbol%3BAcc:HGNC:15553];gene_id=ENSG00000198445;logic_name=ensembl_havana_gene_homo_sapiens;version=4
4 ID=gene:ENSG00000172967;Name=XKR3;biotype=protein_coding;description=XK related 3 [Source:HGNC Symbol%3BAcc:HGNC:28778];gene_id=ENSG00000172967;logic_name=ensembl_havana_gene_homo_sapiens;version=7
5 ID=gene:ENSG00000215568;Name=GAB4;biotype=protein_coding;description=GRB2 associated binding protein family member 4 [Source:HGNC Symbol%3BAcc:HGNC:18325];gene_id=ENSG00000215568;logic_name=ensembl_havana_gene_homo_sapiens;version=9
6 ID=gene:ENSG00000177663;Name=IL17RA;biotype=protein_coding;description=interleukin 17 receptor A [Source:HGNC Symbol%3BAcc:HGNC:5985];gene_id=ENSG00000177663;logic_name=ensembl_havana_gene_homo_sapiens;version=14Estimated splatPopParams
Load 10x-Neuro splatPopParams as estimated here.
params <- readRDS("output/01_sims/splatPop-params_Neuro-10x.rds")
params@eqtl.coreg <- 0
params.emp <- setParams(params, batchCells=100,
eqtl.maf.min=0.05, eqtl.maf.max=1)Additional input data needed
To generate a simulated dataset that has real eQTL effects and real gene means and gene variances, we will also need the aggregated gene means across all cells for each individual for each gene and eQTL mapping results for the genes and variants in the GFF and VCF data. For this demonstration we mapped eQTL using MatrixEQTL (see code/10x_eqtl-mapping-matrixeQTL.R).
Aggregated expression data
sce <- readRDS("data/sce_Neuro-10x_DA_allGenes.rds")
sce$Sample <- gsub("_.*", "", gsub(".*-", "", sce$Sample))
keepSamples <- intersect(unique(sce$Sample), colnames(vcf))
sce <- subset(sce, , Sample %in% keepSamples)
message("Number of samples with genotype & pheotype data: ", length(keepSamples))Number of samples with genotype & pheotype data: 37sce <- sce[row.names(sce) %in% geneKey$symbol, ]
rowData(sce) <- geneKey[row.names(sce), ]
row.names(sce) <- rowData(sce)$feature_id
agg <- read.table("output/10x_eQTL-mapping/expression.tsv", sep="\t", header=TRUE)
message("Number of genes with phenotype data on chr22: ", nrow(agg))Number of genes with phenotype data on chr22: 358sce <- sce[row.names(agg), ]
agg[1:5, 1:5] bubh ceik cuhk deyz eipl
ENSG00000177663 0.00000000 0.00000000 0.001779359 0.003047232 0.00000000
ENSG00000099954 0.02631579 0.01515152 0.008896797 0.025901473 0.00000000
ENSG00000182902 0.00000000 0.00000000 0.000000000 0.001015744 0.01960784
ENSG00000131100 0.68421053 0.60606061 0.523131673 0.513458608 0.58823529
ENSG00000099968 0.05263158 0.06060606 0.049822064 0.071609954 0.05882353Filter GFF and VCF to keep only genes and samples, respectively, that have phenotype data.
gff <- gff[geneKey$feature_id %in% row.names(agg), ]
vcf <- vcf[, keepSamples]
vcf <- vcf[, !duplicated(colnames(vcf))]
maf <- VariantAnnotation::snpSummary(vcf)$a1Freq
maf[is.na(maf)] <- 0
names(maf) <- rownames(vcf)
message("SNPs with MAF > 0.05 and < 0.5: ", nrow(vcf))SNPs with MAF > 0.05 and < 0.5: 4690sce <- subset(sce, , Sample %in% keepSamples)eQTL data
eqtl <- readRDS("output/10x_eQTL-mapping/cis_eqtl_results.rds")
vcf.df <- as.data.frame(rowRanges(vcf))
vcf.df$snpMAF <- VariantAnnotation::snpSummary(vcf)$a1Freq
vcf.df$snps <- row.names(vcf.df)
eqtl2 <- eqtl %>% left_join(vcf.df, by="snps") %>%
dplyr::select(snpID=snps, snpCHR=seqnames, snpLOC=start, geneID=gene,
slope=beta, snpMAF)
missG <- data.frame(geneID=row.names(agg)[!row.names(agg) %in% eqtl2$geneID])
eqtl2 <- plyr::rbind.fill(eqtl2, missG)Example simulations
1. Replicating real population level properties
To achieve this level of empirical data replication, provide splatPopSimulate with the VCF, GFF, splatPopParams, and with a splatPop key. The key can be manually created or we can generate a key from empirical data using splatPopSimulateMeans (shown here).
if(rerun){
simMeans <- splatPopSimulateMeans(vcf = vcf, gff = gff, eqtl = eqtl2,
means = as.matrix(agg), params = params.emp)
simPopu <- splatPopSimulate(vcf = vcf, gff = gff, params = params.emp,
key = simMeans$key, sparsify = FALSE)
if(save){
save.name <- paste0(date, "_10x-Neuro-simMeans")
saveRDS(simMeans, paste0("output/01_sims/", save.name, ".rds"))
save.name <- paste0(date, "_10x-Neuro-simPopu")
saveRDS(simPopu, paste0("output/01_sims/", save.name, ".rds"))
}
} else{
save.name <- paste0(date.use, "_10x-Neuro-simMeans")
simMeans <- readRDS(paste0("output/01_sims/", save.name, ".rds"))
save.name <- paste0(date.use, "_10x-Neuro-simPopu")
simPopu <- readRDS(paste0("output/01_sims/", save.name, ".rds"))
}2. Replicating real individual level properties
To achieve this level of empirical data replication, provide splatPopSimulate with the VCF, GFF, splatPopParams, and with real gene means and eQTL summary statistics (see above for examples).
if(rerun){
simIndiv <- splatPopSimulate(vcf = vcf, gff = gff, eqtl = eqtl2,
means = as.matrix(agg), params = params.emp,
sparsify = FALSE)
if(save){
save.name <- paste0(date, "_10x-Neuro-simIndiv")
saveRDS(simIndiv, paste0("output/01_sims/", save.name, ".rds"))
}
} else{
save.name <- paste0(date.use, "_10x-Neuro-simIndiv")
simIndiv <- readRDS(paste0("output/01_sims/", save.name, ".rds"))
}Plot correlation between individuals
The real relationship between individuals can be visualized using a kinship matrix and using the Pearson’s correlation between pseudo-bulked log normalized expression counts between individuals.
pool <- unique(subset(sce, , Batch == "pool9")$Sample)
kinship <- read.table("data/iPSC_eqtl-mapping/hipsci.chr22.rel_mod")
names(kinship) <- gsub(".*\\.", "", names(kinship))
row.names(kinship) <- gsub(".*-", "", row.names(kinship))
kinshipPool <- kinship[pool, pool]
sce2plot <- subset(sce, , Sample %in% pool)
sce2plot <- logNormCounts(sce2plot)
sceAgg2plot <- aggregateAcrossCells(sce2plot, ids = sce2plot$Sample,
statistics="mean",
use.assay.type="logcounts")
sceCorr <- cor(logcounts(sceAgg2plot), method = c("pearson"))
htSCE <- Heatmap(sceCorr, name = "Empirical", col=col_fun)
col_order = column_order(htSCE)
htKin <- Heatmap(kinshipPool, name = "Kinship", column_order = col_order,
col=circlize::colorRamp2(c(-0.1, 0, 0.1), c("blue", "white", "red")))
htSCE + htKin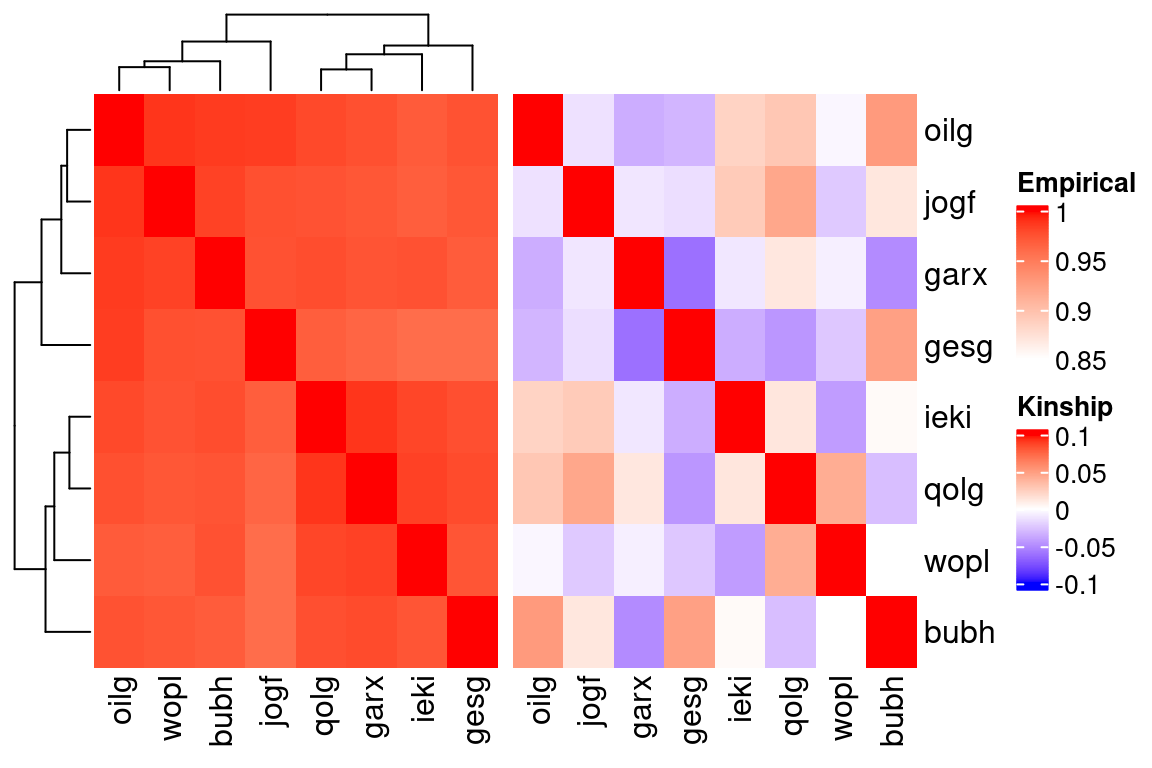
Inter-individual relationships from empirical data. (a) Pearson’s correlation between log normalized mean-aggregated counts and (b) the genetic kinship matrix.
| Version | Author | Date |
|---|---|---|
| 7f8fd63 | cazodi | 2021-09-27 |
simPopu2plot <- subset(simPopu, , Sample %in% pool)
simIndiv2plot <- subset(simIndiv, , Sample %in% pool)
simPopu2plot <- logNormCounts(simPopu2plot)
simIndiv2plot <- logNormCounts(simIndiv2plot)
simPopuAgg2plot <- aggregateAcrossCells(simPopu2plot, ids = simPopu2plot$Sample,
statistics="mean",
use.assay.type="logcounts")
simIndivAgg2plot <- aggregateAcrossCells(simIndiv2plot, ids = simIndiv2plot$Sample,
statistics="mean",
use.assay.type="logcounts")
simPopuCorr <- cor(logcounts(simPopuAgg2plot), method = "pearson")
simIndivCorr <- cor(logcounts(simIndivAgg2plot), method = "pearson")
ht1 = Heatmap(simPopuCorr, name = "Popu", column_order = col_order, col=col_fun)
ht2 = Heatmap(simIndivCorr, name = "Indiv", column_order = col_order, col=col_fun)
htSCE + ht2 + ht1 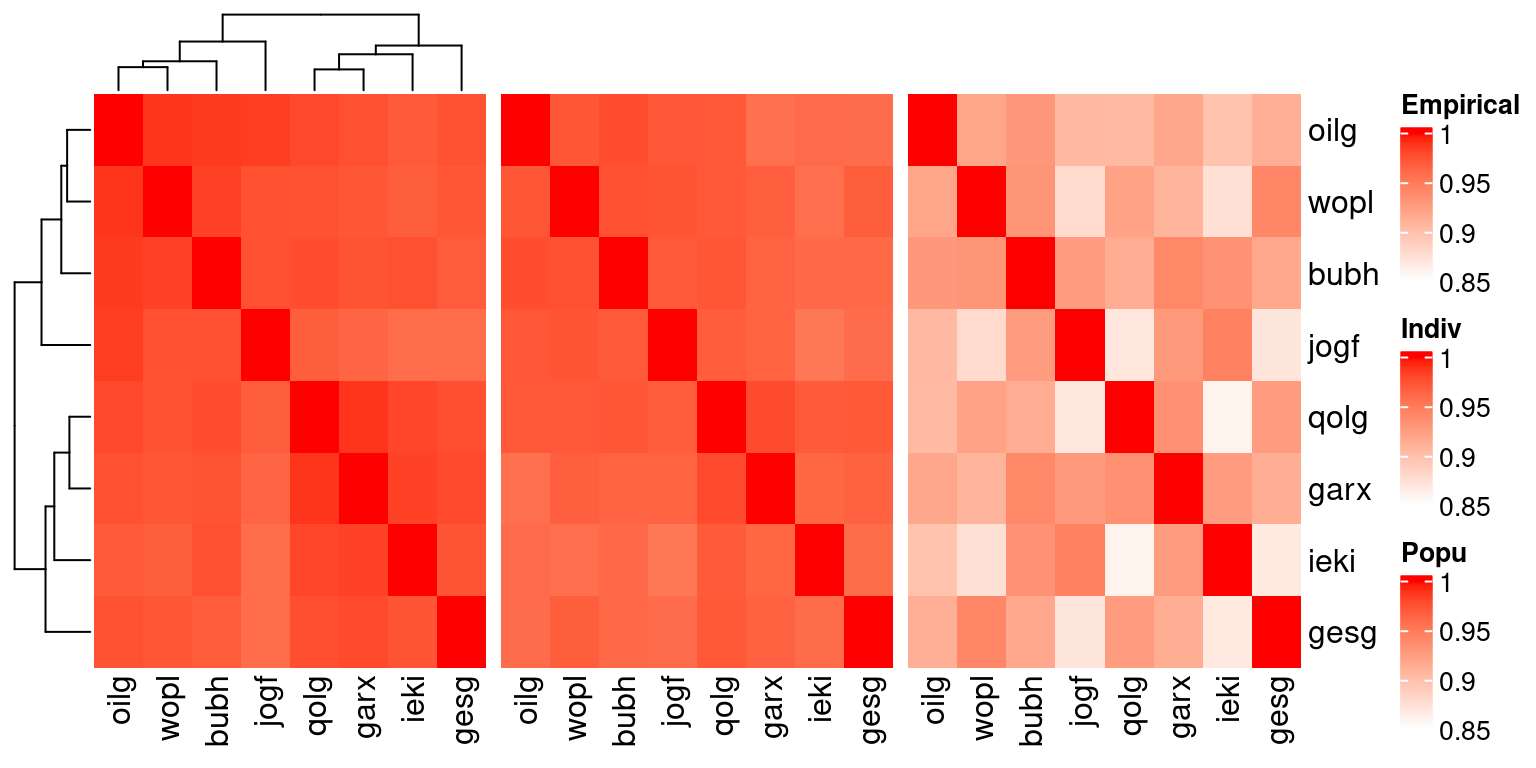
Inter-individual relationships from (left) empirical data, (middle) simulated data replicating individual-level properties, (right) simulated data replicating population-level properties.
| Version | Author | Date |
|---|---|---|
| 7f8fd63 | cazodi | 2021-09-27 |
if(save){
col_fun2 <- circlize::colorRamp2(c(0.9, 1), c("white", "red"))
sceCorrLower <- sceCorr[col_order, col_order]
sceCorrLower[upper.tri(sceCorrLower)] <- 0
simIndivCorrLower <- simIndivCorr[col_order, col_order]
simIndivCorrLower[upper.tri(simIndivCorrLower)] <- 0
htSCE <- Heatmap(sceCorrLower, name = "Empirical", col=col_fun2)
ht2 <- Heatmap(simIndivCorrLower, name = "Indiv", col=col_fun2)
save.name <- paste0("output/00_Figures/", date, "_10x-Neuro_replicate-correlation.pdf")
pdf(save.name, width=6, height=3)
htSCE + ht2
dev.off()
}sceCorrlong <- as.data.frame(sceCorr) %>% rownames_to_column(var="donor1") %>%
pivot_longer(-donor1, names_to = "donor2", values_to = "r")
simIndivCorrlong <- as.data.frame(simIndivCorr) %>%
rownames_to_column(var="donor1") %>%
pivot_longer(-donor1, names_to = "donor2", values_to = "r")
corrPairs <- sceCorrlong %>%
left_join(simIndivCorrlong, by=c("donor1", "donor2"),
suffix = c(".empirical", ".simulated"))
ggscatter(corrPairs, x="r.empirical", y="r.simulated", add = "reg.line",
add.params = list(color = "blue", fill = "lightgray"),
conf.int = TRUE, cor.coef = TRUE, xlim=c(0.95, 1), ylim=c(0.95, 1),
cor.coeff.args = list(method = "pearson", label.x = 0.95, label.sep = "\n"))+
geom_abline()`geom_smooth()` using formula 'y ~ x'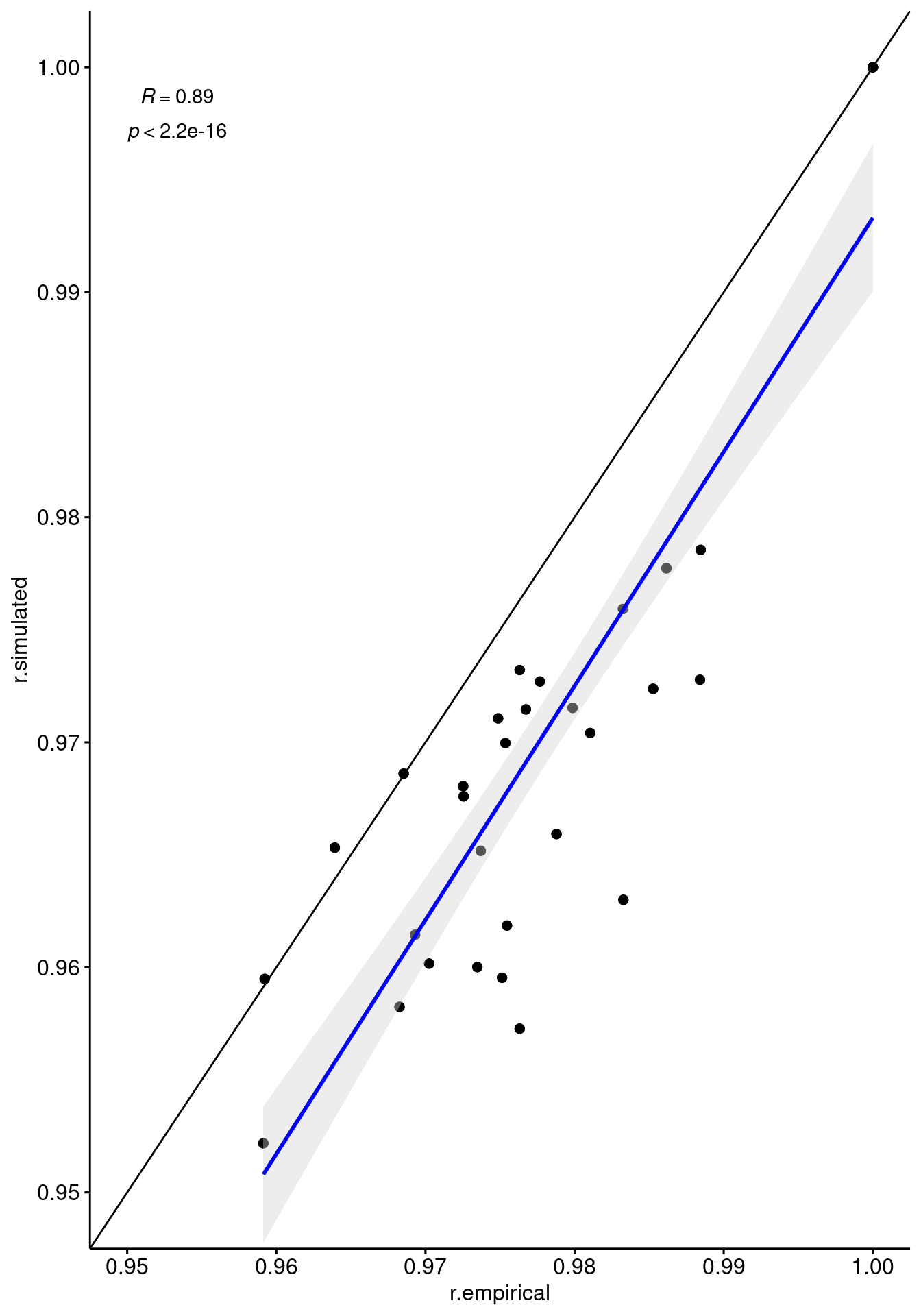
Comparison between the pairwise correlation in expression between pairs of individuals in the empirical (x-axis) and simulated (y-axis) data.
| Version | Author | Date |
|---|---|---|
| 0d55650 | cazodi | 2021-11-19 |
if(save){
save.name <- paste0(date, "_10x-Neuro_replicate-correlation-scatter.pdf")
ggsave(paste0("output/00_Figures/", save.name), width = 4, height = 4)
}Plot patterns of gene expression
top35 <- names(sort(rowMeans(logcounts(sceAgg2plot)), decreasing = TRUE))[1:35]
col_fun3 <- circlize::colorRamp2(c(0, 5), c("black", "green"))
ghSCE <- Heatmap(logcounts(sceAgg2plot)[top35, ], col=col_fun3,
name = "Empirical")
col_order = column_order(ghSCE)
row_order = row_order(ghSCE)
ghSIM <- Heatmap(logcounts(simIndivAgg2plot)[top35, ], name = "Simulated",
column_order = col_order, row_order=row_order, col=col_fun3)
if(save){
save.name <- paste0("output/00_Figures/", date, "_10x-Neuro_replicate-GeneCorrelation.pdf")
pdf(save.name, width=5, height=8)
ghSCE + ghSIM
dev.off()
} else{
ghSCE + ghSIM
}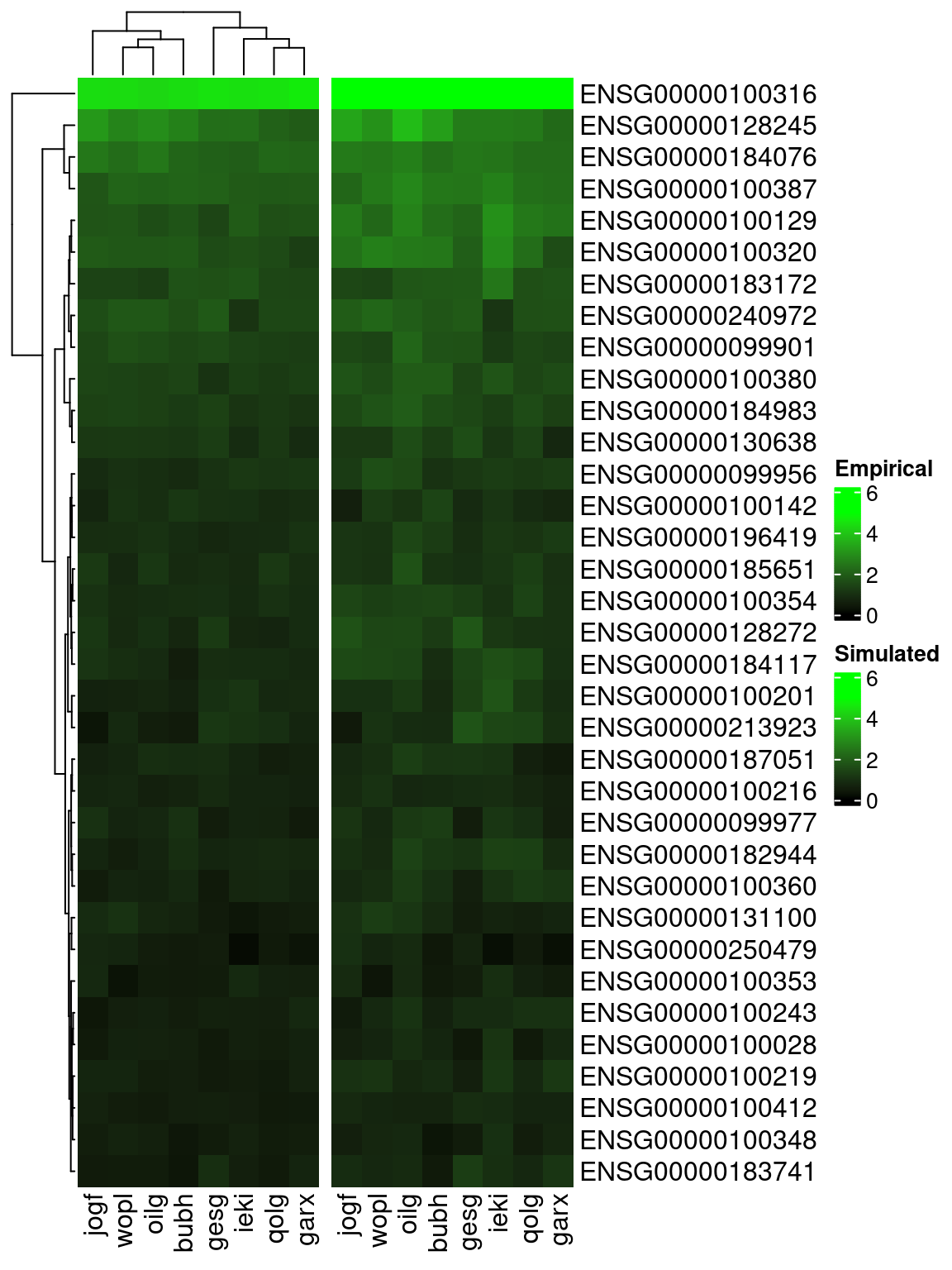
Pattern of mean aggregated logcount expression for top 35 most highly expressed genes (rows) across 8 individuals (columns), from (left) empirical data and (right) splatPop simulated data with individual-level properties replicated.
| Version | Author | Date |
|---|---|---|
| 7f8fd63 | cazodi | 2021-09-27 |
sceLogClong <- as.data.frame(logcounts(sceAgg2plot)) %>%
rownames_to_column(var="gene") %>%
pivot_longer(-gene, names_to = "donor", values_to = "logcount")
simLogClong <- as.data.frame(logcounts(simIndivAgg2plot)) %>%
rownames_to_column(var="gene") %>%
pivot_longer(-gene, names_to = "donor", values_to = "logcount")
logCPairs <- sceLogClong %>%
left_join(simLogClong, by=c("gene", "donor"),
suffix = c(".empirical", ".simulated"))
ggscatter(logCPairs, x="logcount.empirical", y="logcount.simulated",
color="donor", palette = sample.colors,
add = "reg.line", alpha=0.25,
add.params = list(color = "blue", fill = "lightgray"),
conf.int = TRUE, cor.coef = TRUE, xlim = c(0, 7), ylim=c(0,7),
cor.coeff.args = list(method = "pearson", label.x = 0.96, label.sep = "\n")) +
geom_abline()`geom_smooth()` using formula 'y ~ x'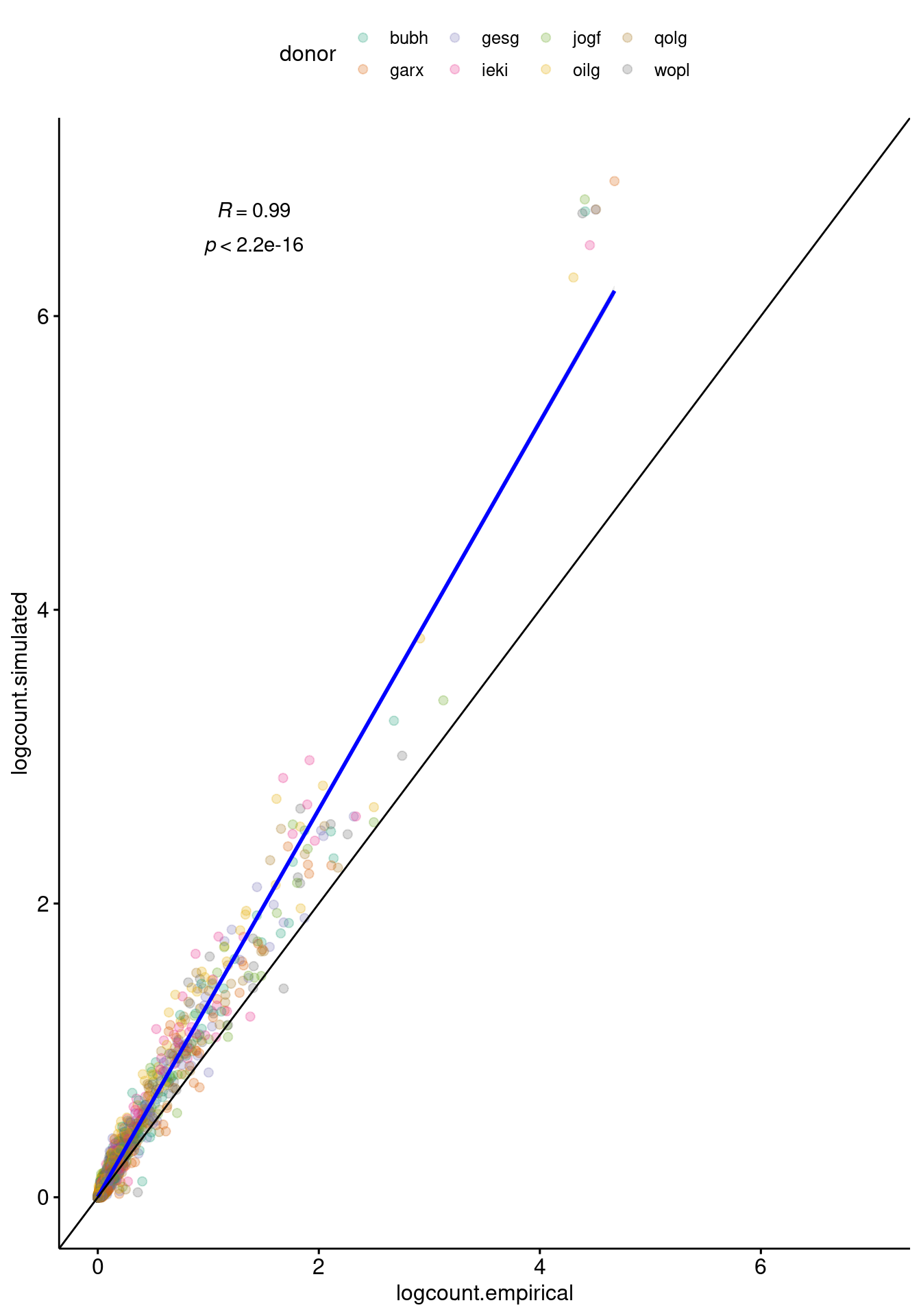
Comparison between the pairwise correlation in expression between pairs of individuals in the empirical (x-axis) and simulated (y-axis) data.
| Version | Author | Date |
|---|---|---|
| 0d55650 | cazodi | 2021-11-19 |
if(save){
save.name <- paste0(date, "_10x-Neuro_gene-logcount-scatter.pdf")
ggsave(paste0("output/00_Figures/", save.name), width = 4, height = 4)
}
devtools::session_info()─ Session info ──────────────────────────────────────────────────────────────
hash: dna, flag: Greece, flag: Thailand
setting value
version R version 4.1.1 (2021-08-10)
os Rocky Linux 8.4 (Green Obsidian)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-11-19
pandoc 2.11.4 @ /usr/lib/rstudio-server/bin/pandoc/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.1.1)
AnnotationDbi 1.56.2 2021-11-09 [1] Bioconductor
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.1.1)
backports 1.3.0 2021-10-27 [1] CRAN (R 4.1.1)
beachmat 2.10.0 2021-10-26 [1] Bioconductor
beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.1.1)
Biobase * 2.54.0 2021-10-26 [1] Bioconductor
BiocFileCache 2.2.0 2021-10-26 [1] Bioconductor
BiocGenerics * 0.40.0 2021-10-26 [1] Bioconductor
BiocIO 1.4.0 2021-10-26 [1] Bioconductor
BiocNeighbors 1.12.0 2021-10-26 [1] Bioconductor
BiocParallel 1.28.0 2021-10-26 [1] Bioconductor
BiocSingular 1.10.0 2021-10-26 [1] Bioconductor
biomaRt 2.50.0 2021-10-26 [1] Bioconductor
Biostrings * 2.62.0 2021-10-26 [1] Bioconductor
bit 4.0.4 2020-08-04 [1] CRAN (R 4.1.1)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.1.1)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.1.1)
blob 1.2.2 2021-07-23 [1] CRAN (R 4.1.1)
broom 0.7.10 2021-10-31 [1] CRAN (R 4.1.1)
BSgenome 1.62.0 2021-10-26 [1] Bioconductor
bslib 0.3.1 2021-10-06 [1] CRAN (R 4.1.1)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.4)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.1)
car 3.0-12 2021-11-06 [1] CRAN (R 4.1.1)
carData 3.0-4 2020-05-22 [1] CRAN (R 4.1.1)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.1.1)
checkmate 2.0.0 2020-02-06 [1] CRAN (R 4.1.1)
circlize 0.4.13 2021-06-09 [1] CRAN (R 4.1.1)
cli 3.1.0 2021-10-27 [1] CRAN (R 4.1.1)
clue 0.3-60 2021-10-11 [1] CRAN (R 4.1.1)
cluster * 2.1.2 2021-04-17 [2] CRAN (R 4.1.1)
codetools 0.2-18 2020-11-04 [1] CRAN (R 4.1.1)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.1.1)
ComplexHeatmap * 2.10.0 2021-10-26 [1] Bioconductor
crayon 1.4.2 2021-10-29 [1] CRAN (R 4.1.1)
curl 4.3.2 2021-06-23 [1] CRAN (R 4.1.1)
data.table * 1.14.2 2021-09-27 [1] CRAN (R 4.0.4)
DBI 1.1.1 2021-01-15 [1] CRAN (R 4.1.1)
dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.1.1)
DelayedArray 0.20.0 2021-10-26 [1] Bioconductor
DelayedMatrixStats 1.16.0 2021-10-26 [1] Bioconductor
desc 1.4.0 2021-09-28 [1] CRAN (R 4.1.1)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.1.1)
digest 0.6.28 2021-09-23 [1] CRAN (R 4.0.4)
doParallel 1.0.16 2020-10-16 [1] CRAN (R 4.1.1)
dplyr * 1.0.7 2021-06-18 [1] CRAN (R 4.1.1)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.1)
evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.1)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.1.1)
farver 2.1.0 2021-02-28 [1] CRAN (R 4.1.1)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.1)
filelock 1.0.2 2018-10-05 [1] CRAN (R 4.1.1)
fitdistrplus * 1.1-6 2021-09-28 [1] CRAN (R 4.1.1)
forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.1.1)
foreach 1.5.1 2020-10-15 [1] CRAN (R 4.1.1)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.1.1)
generics 0.1.1 2021-10-25 [1] CRAN (R 4.1.1)
GenomeInfoDb * 1.30.0 2021-10-26 [1] Bioconductor
GenomeInfoDbData 1.2.7 2021-11-03 [1] Bioconductor
GenomicAlignments 1.30.0 2021-10-26 [1] Bioconductor
GenomicFeatures 1.46.1 2021-10-27 [1] Bioconductor
GenomicRanges * 1.46.0 2021-10-26 [1] Bioconductor
GetoptLong 1.0.5 2020-12-15 [1] CRAN (R 4.1.1)
ggbeeswarm 0.6.0 2017-08-07 [1] CRAN (R 4.1.1)
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.1.1)
ggpubr * 0.4.0 2020-06-27 [1] CRAN (R 4.1.1)
ggrepel 0.9.1 2021-01-15 [1] CRAN (R 4.1.1)
ggsignif 0.6.3 2021-09-09 [1] CRAN (R 4.1.1)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.1.1)
GlobalOptions 0.1.2 2020-06-10 [1] CRAN (R 4.1.1)
glue 1.5.0 2021-11-07 [1] CRAN (R 4.1.1)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.1.1)
gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.1)
haven 2.4.3 2021-08-04 [1] CRAN (R 4.1.1)
highr 0.9 2021-04-16 [1] CRAN (R 4.1.1)
hms 1.1.1 2021-09-26 [1] CRAN (R 4.1.1)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.0.4)
httpuv 1.6.3 2021-09-09 [1] CRAN (R 4.1.1)
httr 1.4.2 2020-07-20 [1] CRAN (R 4.1.1)
IRanges * 2.28.0 2021-10-26 [1] Bioconductor
irlba 2.3.3 2019-02-05 [1] CRAN (R 4.1.1)
iterators 1.0.13 2020-10-15 [1] CRAN (R 4.1.1)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.1.1)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.1)
KEGGREST 1.34.0 2021-10-26 [1] Bioconductor
knitr 1.36 2021-09-29 [1] CRAN (R 4.1.1)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.1.1)
later 1.3.0 2021-08-18 [1] CRAN (R 4.1.1)
lattice 0.20-45 2021-09-22 [1] CRAN (R 4.1.1)
lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.1.1)
locfit 1.5-9.4 2020-03-25 [1] CRAN (R 4.1.1)
lubridate 1.8.0 2021-10-07 [1] CRAN (R 4.0.4)
magick 2.7.3 2021-08-18 [1] CRAN (R 4.1.1)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.1)
MASS * 7.3-54 2021-05-03 [1] CRAN (R 4.1.1)
Matrix 1.3-4 2021-06-01 [1] CRAN (R 4.1.1)
MatrixGenerics * 1.6.0 2021-10-26 [1] Bioconductor
matrixStats * 0.61.0 2021-09-17 [1] CRAN (R 4.1.1)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.1.1)
mgcv 1.8-38 2021-10-06 [1] CRAN (R 4.1.1)
modelr 0.1.8 2020-05-19 [1] CRAN (R 4.1.1)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.1)
nlme 3.1-153 2021-09-07 [1] CRAN (R 4.1.1)
pillar 1.6.4 2021-10-18 [1] CRAN (R 4.1.1)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.1.1)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.1)
pkgload 1.2.3 2021-10-13 [1] CRAN (R 4.1.1)
plyr 1.8.6 2020-03-03 [1] CRAN (R 4.1.1)
png 0.1-7 2013-12-03 [1] CRAN (R 4.1.1)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.1)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.1)
progress 1.2.2 2019-05-16 [1] CRAN (R 4.1.1)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.1.1)
ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.1)
purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.1.1)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.1)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.1.1)
RColorBrewer * 1.1-2 2014-12-07 [1] CRAN (R 4.1.1)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.1.1)
RCurl 1.98-1.5 2021-09-17 [1] CRAN (R 4.1.1)
readr * 2.1.0 2021-11-11 [1] CRAN (R 4.1.1)
readxl 1.3.1 2019-03-13 [1] CRAN (R 4.1.1)
remotes 2.4.1 2021-09-29 [1] CRAN (R 4.1.1)
reprex 2.0.1 2021-08-05 [1] CRAN (R 4.1.1)
restfulr 0.0.13 2017-08-06 [1] CRAN (R 4.1.1)
rjson 0.2.20 2018-06-08 [1] CRAN (R 4.1.1)
rlang 0.4.12 2021-10-18 [1] CRAN (R 4.0.4)
rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.1.1)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.1)
Rsamtools * 2.10.0 2021-10-26 [1] Bioconductor
RSQLite 2.2.8 2021-08-21 [1] CRAN (R 4.0.4)
rstatix 0.7.0 2021-02-13 [1] CRAN (R 4.1.1)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.1)
rsvd 1.0.5 2021-04-16 [1] CRAN (R 4.1.1)
rtracklayer 1.54.0 2021-10-26 [1] Bioconductor
rvest 1.0.2 2021-10-16 [1] CRAN (R 4.1.1)
S4Vectors * 0.32.2 2021-11-07 [1] Bioconductor
sass 0.4.0 2021-05-12 [1] CRAN (R 4.1.1)
ScaledMatrix 1.2.0 2021-10-26 [1] Bioconductor
scales 1.1.1 2020-05-11 [1] CRAN (R 4.1.1)
scater * 1.22.0 2021-10-26 [1] Bioconductor
scuttle * 1.4.0 2021-10-26 [1] Bioconductor
sessioninfo 1.2.1 2021-11-02 [1] CRAN (R 4.1.1)
shape 1.4.6 2021-05-19 [1] CRAN (R 4.1.1)
SingleCellExperiment * 1.16.0 2021-10-26 [1] Bioconductor
sparseMatrixStats 1.6.0 2021-10-26 [1] Bioconductor
splatter * 1.18.1 2021-11-02 [1] Bioconductor
stringi 1.7.5 2021-10-04 [1] CRAN (R 4.0.4)
stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.1.1)
SummarizedExperiment * 1.24.0 2021-10-26 [1] Bioconductor
survival * 3.2-13 2021-08-24 [1] CRAN (R 4.0.4)
testthat 3.1.0 2021-10-04 [1] CRAN (R 4.1.1)
tibble * 3.1.6 2021-11-07 [1] CRAN (R 4.1.1)
tidyr * 1.1.4 2021-09-27 [1] CRAN (R 4.0.4)
tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.1.1)
tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 4.1.1)
tzdb 0.2.0 2021-10-27 [1] CRAN (R 4.1.1)
usethis 2.1.3 2021-10-27 [1] CRAN (R 4.1.1)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.4)
VariantAnnotation * 1.40.0 2021-10-26 [1] Bioconductor
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.1.1)
vipor 0.4.5 2017-03-22 [1] CRAN (R 4.1.1)
viridis 0.6.2 2021-10-13 [1] CRAN (R 4.0.4)
viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.1.1)
whisker 0.4 2019-08-28 [1] CRAN (R 4.1.1)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.1.1)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.1.1)
xfun 0.28 2021-11-04 [1] CRAN (R 4.1.1)
XML 3.99-0.8 2021-09-17 [1] CRAN (R 4.1.1)
xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.2)
XVector * 0.34.0 2021-10-26 [1] Bioconductor
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.1)
zlibbioc 1.40.0 2021-10-26 [1] Bioconductor
[1] /mnt/mcfiles/cazodi/R/x86_64-pc-linux-gnu-library/4.1
[2] /opt/R/4.1.1/lib/R/library
──────────────────────────────────────────────────────────────────────────────