Applications: DEG analysis
Christina B. Azodi
2021-09-21
Last updated: 2021-09-21
Checks: 6 1
Knit directory: KEJP_2020_splatPop/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210215) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f729828. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/10x_lung/
Ignored: data/ALL.chr2.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.EURO.0.99.MAF05.filtered.vcf
Ignored: data/D30.h5
Ignored: data/Diabetes/
Ignored: data/IBD/
Ignored: data/agg_10X-fibroblasts-control.rds
Ignored: data/agg_IBD-ss2.rds
Ignored: data/agg_Neuro-10x_DA.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/agg_NeuroSeq-10x_D11-pool6.rds
Ignored: data/agg_diabetes-ss2.rds
Ignored: data/agg_iPSC-ss2_D0.rds
Ignored: data/covid/
Ignored: data/cuomo_NeuroSeq_10x_all_sce.rds
Ignored: data/iPSC_eqtl-mapping/
Ignored: data/lung_fibrosis/
Ignored: data/pseudoB_IBD-ss2.rds
Ignored: data/pseudoB_Neuro-10x_DA.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/pseudoB_NeuroSeq-10x_D11-pool6.rds
Ignored: data/pseudoB_diabetes-ss2.rds
Ignored: data/pseudoB_iPSC-ss2_D0.rds
Ignored: data/sce_10X-fibroblasts-947170.rds
Ignored: data/sce_10X-fibroblasts-allGenes.rds
Ignored: data/sce_10X-fibroblasts.rds
Ignored: data/sce_IBD-ss2.rds
Ignored: data/sce_IBD-ss2_HC2.rds
Ignored: data/sce_Neuro-10x.rds
Ignored: data/sce_Neuro-10x_2CT.rds
Ignored: data/sce_Neuro-10x_DA-wihj4.rds
Ignored: data/sce_Neuro-10x_DA_allGenes.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-filt.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6-mita1FPP.rds
Ignored: data/sce_NeuroSeq-10x_D11-pool6.rds
Ignored: data/sce_diabetes-ss2.rds
Ignored: data/sce_diabetes-ss2_T2D-5.rds
Ignored: data/sce_iPSC-ss2_D0-allGenes.rds
Ignored: data/sce_iPSC-ss2_D0-joxm39.rds
Ignored: data/sce_iPSC-ss2_D0.rds
Ignored: output/00_Figures/
Ignored: output/01_sims/
Ignored: output/10x_eQTL-mapping/
Ignored: output/demo_eQTL/
Ignored: output/iPSC_eQTL/
Ignored: references/1000GP_Phase3_sample_info.txt
Ignored: references/Homo_sapiens.GRCh38.99.chromosome.22.gff3
Ignored: references/chr2.filt.2-temporary.bed
Ignored: references/chr2.filt.2-temporary.bim
Ignored: references/chr2.filt.2-temporary.fam
Ignored: references/chr2.filt.2.log
Ignored: references/chr2.filt.log
Ignored: references/chr2.filt.map
Ignored: references/chr2.filt.nosex
Ignored: references/chr2.filt.ped
Ignored: references/chr2.filt.prune.in
Ignored: references/chr2.filt.prune.out
Ignored: references/chr2.filtered.log
Ignored: references/chr2.filtered.nosex
Ignored: references/chr2.filtered.vcf
Ignored: references/chr2.filtered2.log
Ignored: references/chr2.filtered2.nosex
Ignored: references/chr2.filtered2.vcf
Ignored: references/chr2.filtered3.log
Ignored: references/chr2.filtered3.nosex
Ignored: references/chr2.filtered3.vcf
Ignored: references/chr2.genes.gff3
Ignored: references/chr2.vcf.gz
Ignored: references/chr22.filt.log
Ignored: references/chr22.filt.map
Ignored: references/chr22.filt.nosex
Ignored: references/chr22.filt.ped
Ignored: references/chr22.filt.prune.in
Ignored: references/chr22.filt.prune.out
Ignored: references/chr22.filtered.log
Ignored: references/chr22.filtered.nosex
Ignored: references/chr22.filtered.vcf
Ignored: references/chr22.filtered.vcf.bed
Ignored: references/chr22.filtered.vcf.bim
Ignored: references/chr22.filtered.vcf.fam
Ignored: references/chr22.filtered.vcf.log
Ignored: references/chr22.filtered.vcf.nosex
Ignored: references/chr22.filtered.vcf.rel
Ignored: references/chr22.filtered.vcf.rel.id
Ignored: references/chr22.filtered.vcf.rel_mod
Ignored: references/chr22.genes.gff3
Ignored: references/chr22.genes.gff3_annotation
Ignored: references/chr22.genes.gff3_chunks
Ignored: references/chr22.vcf.gz
Ignored: references/eqtl_summary_stats.tar.gz
Ignored: references/eqtl_summary_stats_renamed/
Ignored: references/keep_samples.txt
Ignored: references/remove_snps.txt
Ignored: references/test.genome
Ignored: references/test.log
Ignored: references/test.nosex
Untracked files:
Untracked: .DS_Store
Untracked: .cache/
Untracked: .config/
Untracked: .snakemake/
Untracked: KEJP_2020_splatPop.Rproj
Untracked: analysis/10x-Neuro_emp-simulations.Rmd
Untracked: code/10x_eqtl-mapping-matrixeQTL.R
Untracked: code/splatter_1.17.2.tar.gz
Untracked: docs/.DS_Store
Unstaged changes:
Modified: .gitignore
Modified: .gitlab-ci.yml
Modified: CITATION
Modified: Dockerfile
Modified: LICENSE
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/10x-Neuro_estimate-params.Rmd
Modified: analysis/Applications_DEG-analysis.Rmd
Modified: analysis/Applications_eQTL-mapping.Rmd
Modified: analysis/KEJP_iPSC-ss2.Rmd
Modified: analysis/_site.yml
Modified: analysis/about.Rmd
Modified: analysis/index.Rmd
Modified: analysis/license.Rmd
Modified: analysis/misc_analysis-time.Rmd
Modified: analysis/ss2-iPSC_emp-simulation.Rmd
Modified: analysis/ss2-iPSC_estimate-params.Rmd
Modified: analysis/ss2-iPSC_simulations.Rmd
Modified: cluster.json
Modified: code/1_process-empirical-data.R
Modified: code/2_estimate-splatPopParams.R
Modified: code/4_simulate_DEG.R
Modified: code/README.md
Modified: code/plot_functions.R
Modified: code/plot_functions2.R
Modified: data/README.md
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.css
Modified: docs/site_libs/anchor-sections-1.0/anchor-sections.js
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap-theme.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.css.map
Modified: docs/site_libs/bootstrap-3.3.5/css/bootstrap.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cerulean.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/cosmo.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/darkly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/flatly.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Lato.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/LatoItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycle.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/NewsCycleBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSans.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansBoldItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/OpenSansLightItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Raleway.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RalewayBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Roboto.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/RobotoMedium.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansPro.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProBold.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProItalic.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/SourceSansProLight.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/fonts/Ubuntu.ttf
Modified: docs/site_libs/bootstrap-3.3.5/css/journal.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/lumen.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/paper.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/readable.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/sandstone.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/simplex.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/spacelab.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/united.min.css
Modified: docs/site_libs/bootstrap-3.3.5/css/yeti.min.css
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.eot
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.svg
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.ttf
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff
Modified: docs/site_libs/bootstrap-3.3.5/fonts/glyphicons-halflings-regular.woff2
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.js
Modified: docs/site_libs/bootstrap-3.3.5/js/bootstrap.min.js
Modified: docs/site_libs/bootstrap-3.3.5/js/npm.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/html5shiv.min.js
Modified: docs/site_libs/bootstrap-3.3.5/shim/respond.min.js
Modified: docs/site_libs/font-awesome-5.1.0/css/all.css
Modified: docs/site_libs/font-awesome-5.1.0/css/v4-shims.css
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-brands-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-regular-400.woff2
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.eot
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.svg
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.ttf
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff
Modified: docs/site_libs/font-awesome-5.1.0/webfonts/fa-solid-900.woff2
Modified: docs/site_libs/header-attrs-2.5/header-attrs.js
Modified: docs/site_libs/jquery-1.11.3/jquery.min.js
Modified: docs/site_libs/jqueryui-1.11.4/README
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_444444_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_555555_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777620_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_777777_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_cc0000_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/images/ui-icons_ffffff_256x240.png
Modified: docs/site_libs/jqueryui-1.11.4/index.html
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.min.js
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.structure.min.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.css
Modified: docs/site_libs/jqueryui-1.11.4/jquery-ui.theme.min.css
Modified: docs/site_libs/navigation-1.1/codefolding.js
Modified: docs/site_libs/navigation-1.1/sourceembed.js
Modified: docs/site_libs/navigation-1.1/tabsets.js
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.css
Modified: docs/site_libs/tocify-1.9.1/jquery.tocify.js
Modified: environment.yml
Modified: envs/limix_env.yaml
Modified: envs/myenv.yaml
Modified: org/README.md
Modified: org/project_management.org
Modified: output/README.md
Modified: references/README.md
Modified: resources/README.md
Modified: resources/keep_samples.txt
Modified: workflow/config_bp_ss2.yaml
Modified: workflow/sims_limix_v1.15.1.smk
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Applications_DEG-analysis.Rmd) and HTML (public/Applications_DEG-analysis.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 7581c38 | cazodi | 2021-08-31 | added multichemistry batch effect example |
| Rmd | e7a6d33 | cazodi | 2021-06-08 | switch from monocle to wilcox test |
| html | e7a6d33 | cazodi | 2021-06-08 | switch from monocle to wilcox test |
| Rmd | 3f88d86 | cazodi | 2021-06-02 | conditional simulations added |
| html | 3f88d86 | cazodi | 2021-06-02 | conditional simulations added |
suppressPackageStartupMessages({
library(splatter)
library(scater)
library(VariantAnnotation)
library(tidyverse)
library(RColorBrewer)
library(ggpubr)
library(MAST)
})
source("code/plot_functions.R")
source("code/misc_functions.R")date <- Sys.Date()
save <- TRUE
rerun <- FALSE
date.use <- "2021-06-08"
q.thresh <- 0.05
nSamples <- 12Simulated data with conditional effects
Generate ss2-iPSC simulated data with conditional DEG effects. Using the same approach as shown here in detail.
if(rerun){
# Chromosome 22 data
gff <- read.table("references/chr22.genes.gff3", sep="\t", header=FALSE, quote="")
vcf <- readVcf("references/chr22.filtered.vcf", "hg38")
sampleNames <- colnames(geno(vcf)$GT)
vcf8 <- vcf[, sample(sampleNames, nSamples)]
params <- readRDS("output/01_sims/splatPop-params_iPSC-ss2_sc.rds")
paramsDE<- setParams(params, batchCells = 500, condition.prob = c(0.5, 0.5),
cde.prob = 0.4, cde.facLoc = 0.2, cde.facScale = 0.2)
sim <- splatPopSimulate(vcf = vcf8, gff = gff, params = paramsDE, sparsify = FALSE)
colnames(sim) <- paste(sim$Sample, sim$Cell, sep=".")
rowData(sim)$delta <- abs(rowData(sim)$ConditionDE.Condition1 -
rowData(sim)$ConditionDE.Condition2)
rowData(sim)$gene_short_name <- rownames(sim)
if(save){saveRDS(sim, paste0("output/01_sims/", date, "_DEG-sim.rds")) }
} else {
sim <- readRDS(paste0("output/01_sims/", date.use, "_DEG-sim.rds"))
}
hist(rowData(sim)$delta)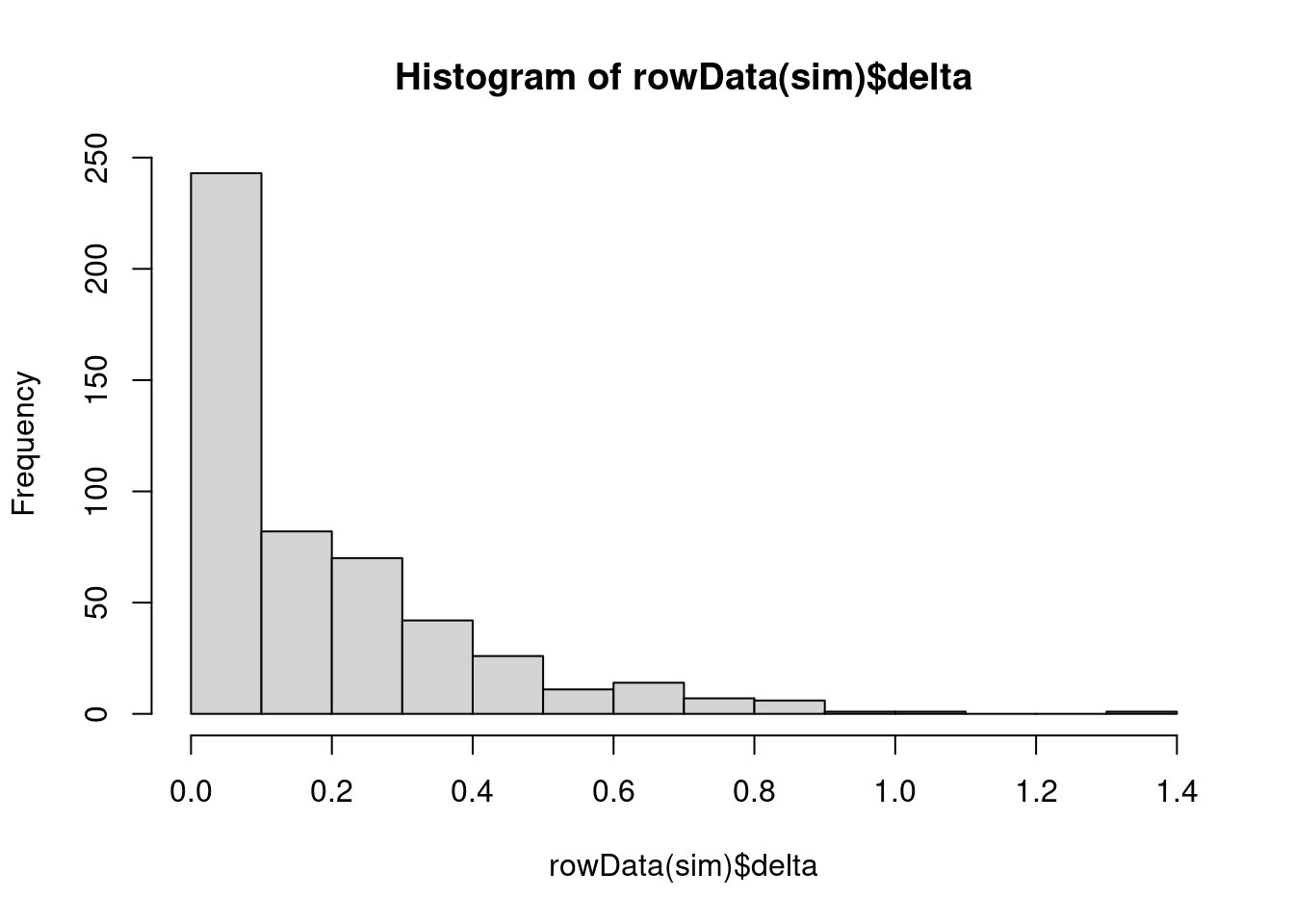
Distribution of DEG effect sizes.
DE analysis: Wilcoxon Rank Sum tests
Results are corrected for multiple testing using Benjamini-Hochberg FDR, with FDR < 0.05 considered significant.
wilcox.Cond2 <- function(agg){
counts_mat <- counts(agg)
lib_size <- colSums(counts_mat)
norm <- t(t(counts_mat)/lib_size * median(lib_size))
group <- colData(agg)["Condition"][colnames(norm), ]
# Run wilcox test
wrt <- as.data.frame(list(p.value = apply(norm, 1, function(x) {
wilcox.test(x[group == "Condition1"], x[group == "Condition2"])$p.value})))
wrt$q.value <- p.adjust(wrt$p.value, method = "fdr")
results <- data.frame(cbind(rowData(agg), wrt))
return (results)
}
summarize.DGE <- function(results, q.thresh){
results$sim <- ifelse(results$delta == 0, 0, 1)
results$est <- ifelse(results$q.value < q.thresh, 1, 0)
results$result <- ifelse(results$sim == 1 & results$est == 1, "TP",
ifelse(results$sim == 0 & results$est == 0, "TN",
ifelse(results$sim == 0 & results$est == 1, "FP",
ifelse(results$sim == 1 & results$est == 0, "FN", NA))))
results$eQTL.EffectSize[results$eQTL.EffectSize == 0] <- NA
results <- as.data.frame(results)
return(results)
}Properties of discovered DEGs
For simulations with 8 samples simulated with four assigned to each conditional group, with 80 cells per sample.
- TP: Simulated DEG, estimated as DEG (q < 0.05)
- FN: Simulated DEG, not significant DEG
- FP: Not simulated as DEG, estimated as DEG
- TN: Not simulated as DEG, not significant DEG
# Down-sample cells
cellsKeep <- paste0("Cell", 1:80)
simSubset <- subset(sim, , Cell %in% cellsKeep)
# Aggregate and run DEG test
sim100_agg <- aggregateAcrossCells(simSubset, ids = simSubset$Sample)
dgt <- wilcox.Cond2(sim100_agg)
dgt <- summarize.DGE(dgt, q.thresh)
table(dgt$result)
FN FP TN TP
267 1 174 62 dgt %>% mutate(mean = meanSampled, CV = cvSampled,
beta = eQTL.EffectSize) %>%
mutate(beta = abs(beta)) %>%
pivot_longer(c("mean", "CV", "delta", "beta"), names_to="factor") %>%
mutate(factor = factor(factor, c("mean", "CV", "beta", "delta"))) %>%
dplyr::filter(result %in% c("TP", "FP", "FN")) %>%
mutate(result = factor(result, c("TP", "FN", "FP"))) %>%
ggboxplot("result", "value", color="result", legend="none",
add="jitter", add.params = list(alpha=0.1),
palette = projectColors("eqtl.result")) +
theme(axis.title.x = element_blank()) +
geom_hline(aes(yintercept=0), linetype="dashed") +
facet_wrap(. ~ factor, scales = "free_y", nrow=1) +
stat_compare_means(aes(label = ..p.signif..),
method = "t.test", ref.group = "TP")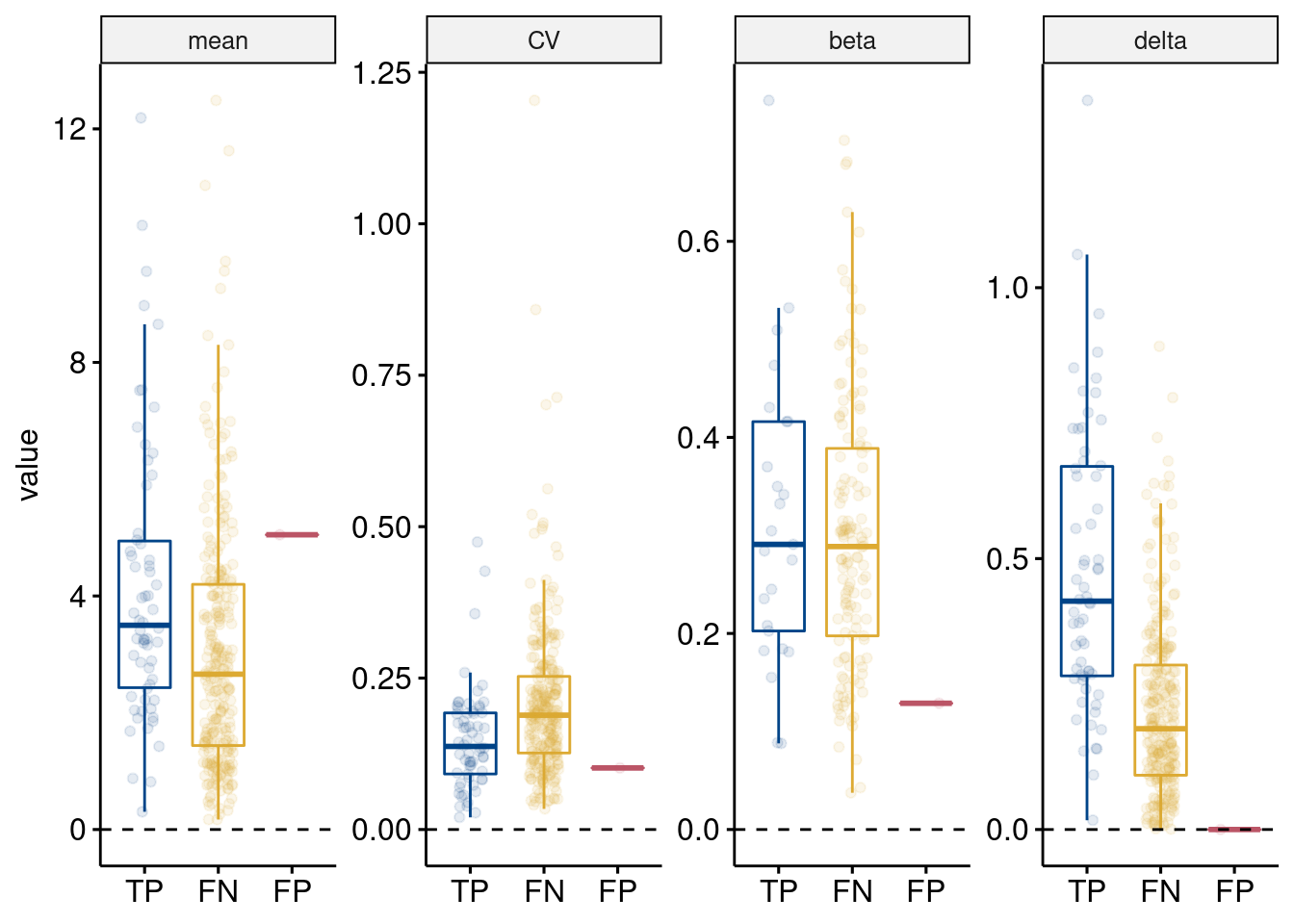
Simulated properties of TP, FN, FP, and TN Wilcoxon rank sum test DE genes. Properties are (left to right) simulated gene mean, gene variance (coefficient of variation), eQTL effect size (beta) if the gene was simulated as an eGene (i.e. zeros, or no eQTL effect, values were removed), and the DE effect size (delta). No delta for FP and TN genes as they were not simulated as DEG. Results from t-tests compared to TP are shown (ns: p > 0.05; \(*\): p <= 0.05; \(**\): p <= 0.01; \(***\): p <= 0.001; \(****\): p <= 0.0001)
if(save){
save.name <- paste0(date, "_ss2-DEG-properties")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 7, height = 3)
}Performance relative to nCells
For simulations with 8 samples simulated with four assigned to each conditional group.
nCellsList <- c(10, 20, 40, 60, 80, 100, 200, 300, 400, 500)
res <- list()
for (i in 1:length(nCellsList)){
nc <- nCellsList[i]
cellsKeep <- paste0("Cell", 1:nc)
tmp <- subset(sim, , Cell %in% cellsKeep)
agg <- aggregateAcrossCells(tmp, ids = tmp$Sample)
dgt <- wilcox.Cond2(agg)
dgt <- summarize.DGE(dgt, q.thresh)
perf <- table(dgt$result)
missing_col <- setdiff(c('TP', 'FN', "TN", 'FP'), names(perf))
perf[missing_col] <- 0
power <- perf[['TP']] / (perf[['TP']] + perf[['FN']])
FDR <- perf[['FP']] / (perf[['FP']] + perf[['TP']])
res[[i]] <- data.frame(list(nCells=nc, TP=perf[['TP']], TN=perf[['TN']],
FP=perf[['FP']], FN=perf[['FN']],
power=power, FDR=FDR))
}
dgtResults <- do.call(rbind, res)
dgtResults %>% pivot_longer(c("power", "FDR"), names_to="metric") %>%
ggline(x = "nCells", y = "value", color = "metric", size=1,
palette = projectColors("eqtl.metrics"), legend="right")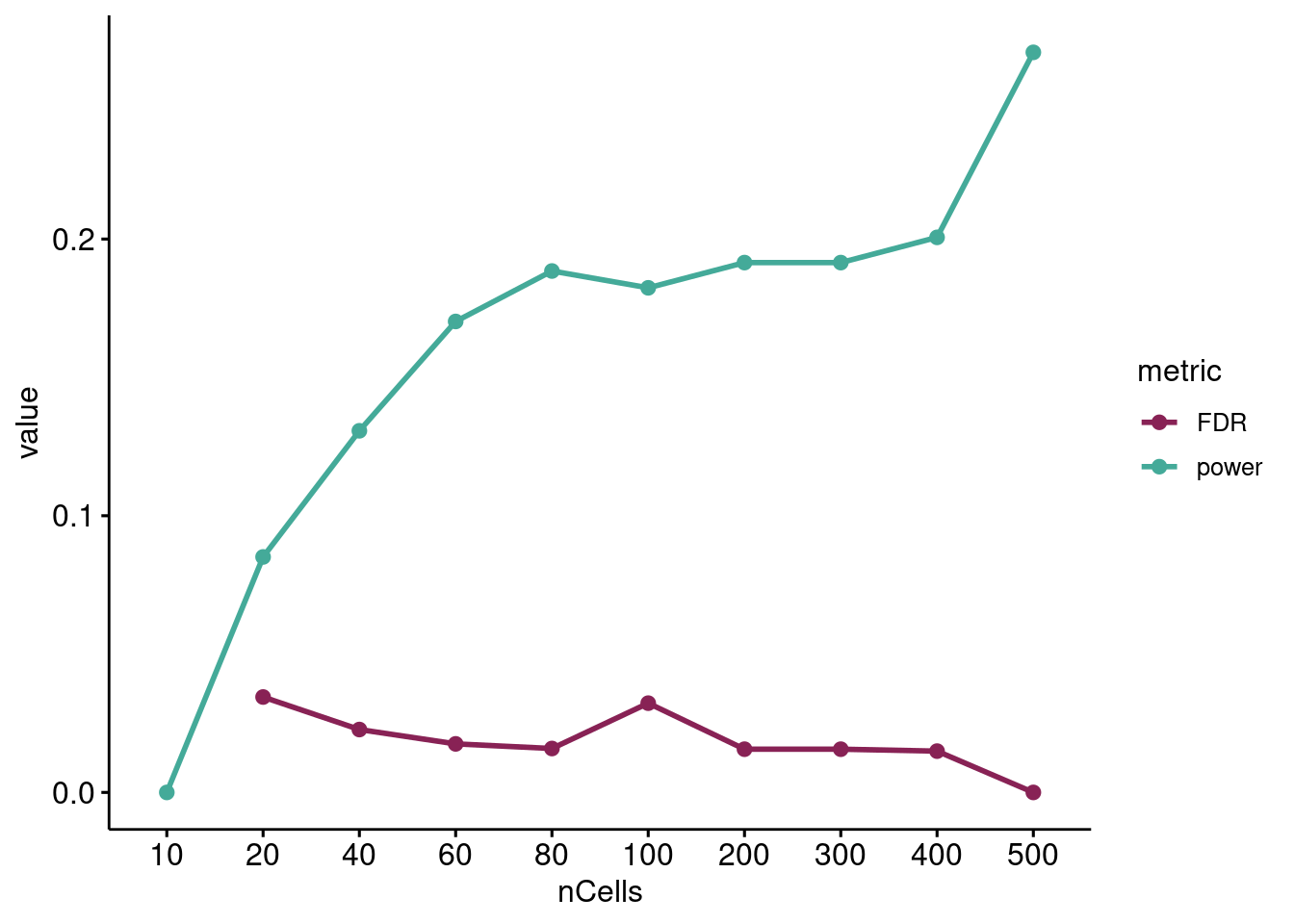
Change in Wilcoxon rank sum test DEG discovery power (TP/TP+FN) and empirical false discovery rate (FP/FP+TP) as the number of cells per sample increases.
if(save){
save.name <- paste0(date, "_ss2-DEG-nCells")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 5, height = 3)
}DE analysis: MAST
MAST was implemented as shown here using zero inflated regression models. Results are corrected for multiple testing using Benjamini-Hochberg FDR, with FDR < 0.05 considered significant.
mast.Cond2 <- function(sim){
sim <- logNormCounts(sim)
sca <- SceToSingleCellAssay(sim)
# run DEG test
zlmCond <- zlm( ~ Condition, sca = sca, exprs_value = 'logcounts')
summaryCond <- summary(zlmCond, doLRT='ConditionCondition2')$datatable
summaryH <- summaryCond[contrast=='ConditionCondition2' & component=='H',.(primerid, `Pr(>Chisq)`)]
summaryFC <- summaryCond[contrast=='ConditionCondition2' & component=='logFC', .(primerid, coef, ci.hi, ci.lo)]
results <- merge(summaryH, summaryFC, by='primerid')
results <- merge(results, as.data.frame(mcols(sca)), by="primerid")
results$q.value <- p.adjust(results$`Pr(>Chisq)`, 'fdr')
return(results)
}Properties of discovered DEGs
For simulations with 8 samples simulated with four assigned to each conditional group, with 80 cells per sample.
- TP: Simulated DEG, estimated as DEG (q < 0.05)
- FN: Simulated DEG, not significant DEG
- FP: Not simulated as DEG, estimated as DEG
- TN: Not simulated as DEG, not significant DEG
dgt <- mast.Cond2(simSubset)
dgt <- summarize.DGE(dgt, q.thresh)
table(dgt$result)
FN FP TN TP
119 52 123 210 dgt %>% mutate(mean = meanSampled, CV = cvSampled,
beta = eQTL.EffectSize) %>%
mutate(beta = abs(beta)) %>%
pivot_longer(c("mean", "CV", "delta", "beta"), names_to="factor") %>%
mutate(factor = factor(factor, c("mean", "CV", "beta", "delta"))) %>%
dplyr::filter(result %in% c("TP", "FP", "FN")) %>%
mutate(result = factor(result, c("TP", "FN", "FP"))) %>%
ggboxplot("result", "value", color="result", legend="none",
add="jitter", add.params = list(alpha=0.1),
palette = projectColors("eqtl.result")) +
theme(axis.title.x = element_blank()) +
geom_hline(aes(yintercept=0), linetype="dashed") +
facet_wrap(. ~ factor, scales = "free_y", nrow=1) +
stat_compare_means(aes(label = ..p.signif..),
method = "t.test", ref.group = "TP")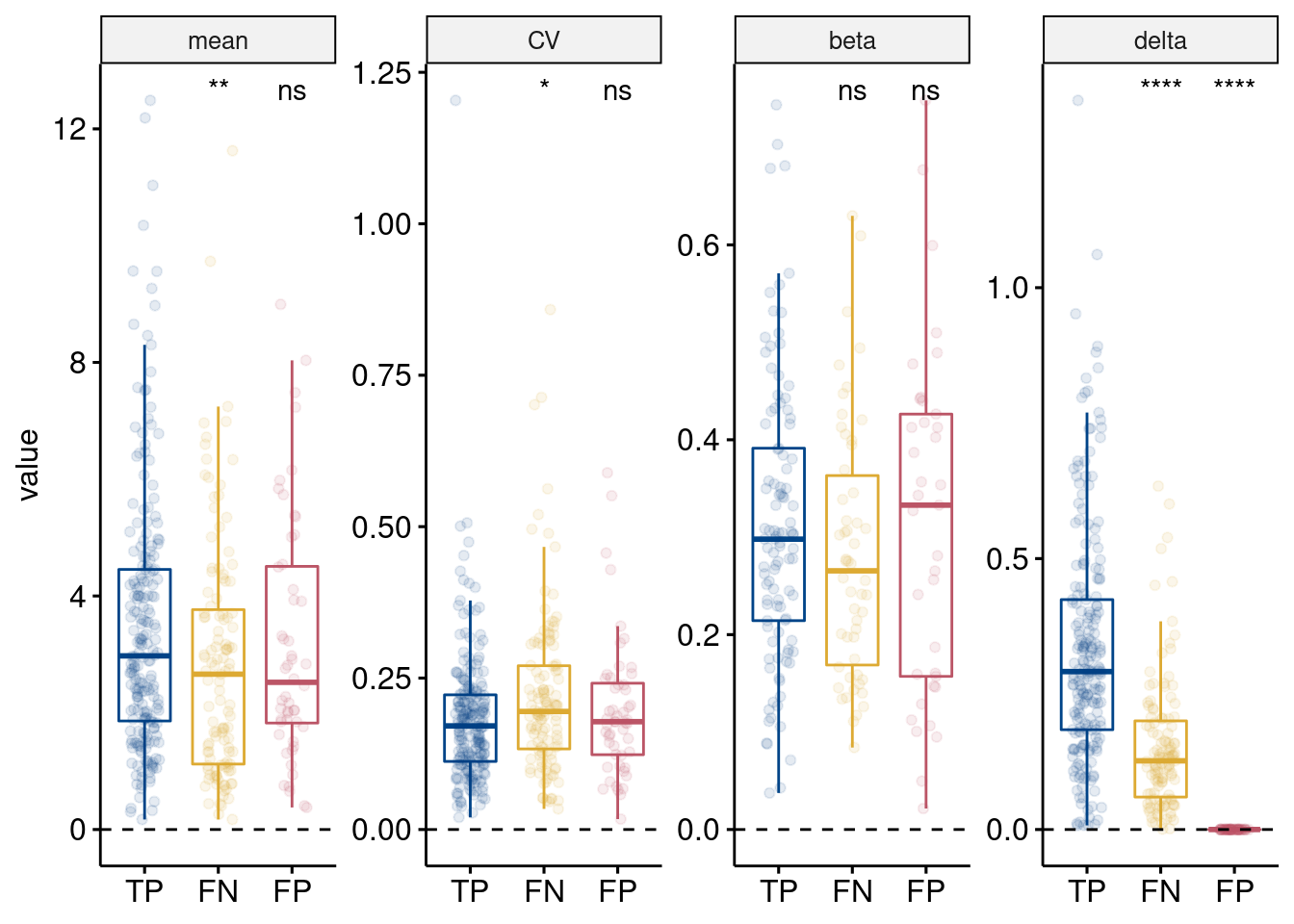
Simulated properties of TP, FN, FP, and TN DE genes from MAST. Properties are (left to right) simulated gene mean, gene variance (coefficient of variation), eQTL effect size (beta) if the gene was simulated as an eGene (i.e. zeros, or no eQTL effect, values were removed), and the DE effect size (delta). No delta for FP and TN genes as they were not simulated as DEG. Results from t-tests compared to TP are shown (ns: p > 0.05; \(*\): p <= 0.05; \(**\): p <= 0.01; \(***\): p <= 0.001; \(****\): p <= 0.0001)
if(save){
save.name <- paste0(date, "_ss2-MAST-DEG-properties")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 7, height = 3)
}Estimated v simulated DE sizes
dgt %>% mutate(coef = abs(coef)) %>%
ggscatter(x="delta", y="coef", color="result",
palette = projectColors("eqtl.result"), alpha = 0.5,
xlab = "simulated delta", ylab = "estimated delta")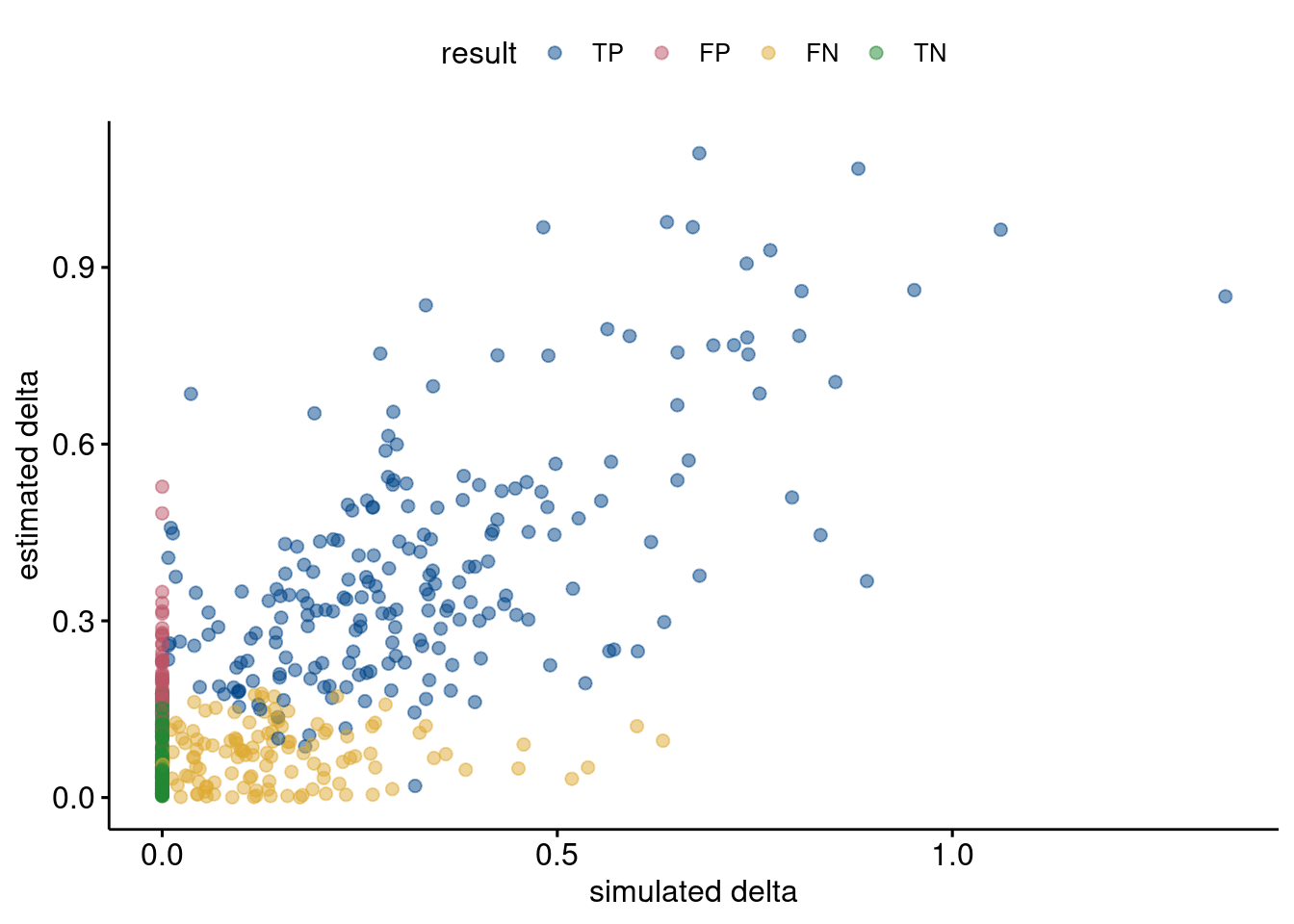
if(save){
save.name <- paste0(date, "_ss2-MAST-DEG-deltas")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 4, height = 4)
}tpCor <- dgt %>% mutate(coef = abs(coef)) %>% filter(result == "TP") %>%
dplyr::summarize(cor(delta, coef))
print("Correlation between estimated and simulated DEG deltas for TPs: ",
round(tpCor$`cor(delta, coef)`, 3))[1] "Correlation between estimated and simulated DEG deltas for TPs: "Performance relative to nCells
nCellsList <- c(10, 20, 40, 60, 80, 100, 200, 300, 400, 500)
res <- list()
for (i in 1:length(nCellsList)){
nc <- nCellsList[i]
cellsKeep <- paste0("Cell", 1:nc)
tmp <- subset(sim, , Cell %in% cellsKeep)
dgt <- mast.Cond2(tmp)
dgt <- summarize.DGE(dgt, q.thresh)
perf <- table(dgt$result)
missing_col <- setdiff(c('TP', 'FN', "TN", 'FP'), names(perf))
perf[missing_col] <- 0
power <- perf[['TP']] / (perf[['TP']] + perf[['FN']])
FDR <- perf[['FP']] / (perf[['FP']] + perf[['TP']])
res[[i]] <- data.frame(list(nCells=nc, TP=perf[['TP']], TN=perf[['TN']],
FP=perf[['FP']], FN=perf[['FN']],
power=power, FDR=FDR))
}
dgtResults <- do.call(rbind, res)
dgtResults %>% pivot_longer(c("power", "FDR"), names_to="metric") %>%
ggline(x = "nCells", y = "value", color = "metric", size=1,
palette = projectColors("eqtl.metrics"), legend="right")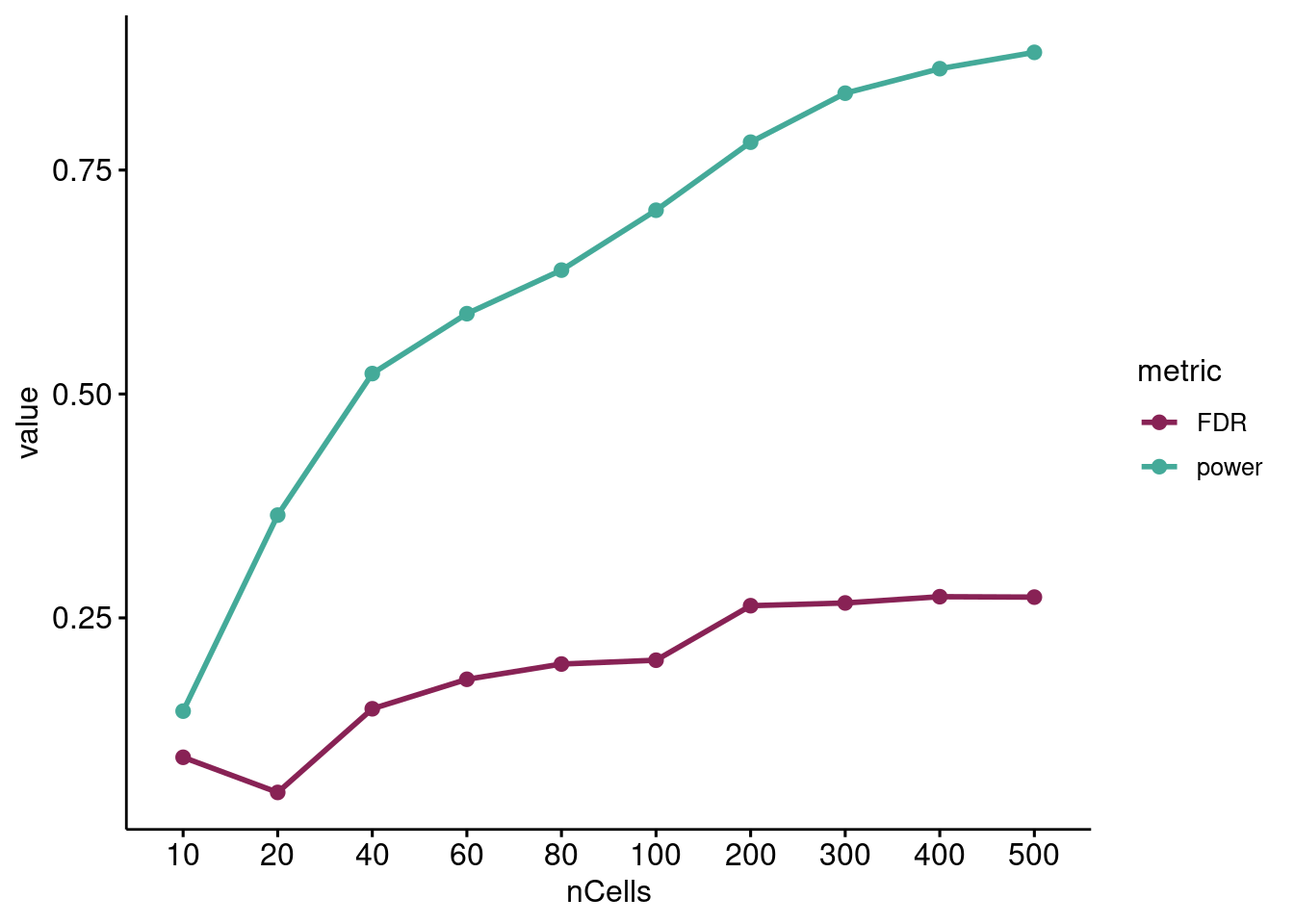
Change in DEG discovery power (TP/TP+FN) and empirical false discovery rate (FP/FP+TP) as the number of cells per sample increases using MAST.
if(save){
save.name <- paste0(date, "_ss2-MAST-DEG-nCells")
ggsave(paste0("output/00_Figures/", save.name, ".pdf"), width = 5, height = 3)
}
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.0.4 (2021-02-15)
os Red Hat Enterprise Linux 8.4 (Ootpa)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2021-09-21
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.0.2)
AnnotationDbi 1.52.0 2020-10-27 [1] Bioconductor
askpass 1.1 2019-01-13 [1] CRAN (R 4.0.2)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.2)
backports 1.2.1 2020-12-09 [1] CRAN (R 4.0.4)
beachmat 2.6.4 2020-12-20 [1] Bioconductor
beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.0.4)
Biobase * 2.50.0 2020-10-27 [1] Bioconductor
BiocFileCache 1.14.0 2020-10-27 [1] Bioconductor
BiocGenerics * 0.36.1 2021-04-16 [1] Bioconductor
BiocNeighbors 1.8.2 2020-12-07 [1] Bioconductor
BiocParallel 1.24.1 2020-11-06 [1] Bioconductor
BiocSingular 1.6.0 2020-10-27 [1] Bioconductor
biomaRt 2.46.3 2021-02-09 [1] Bioconductor
Biostrings * 2.58.0 2020-10-27 [1] Bioconductor
bit 4.0.4 2020-08-04 [1] CRAN (R 4.0.2)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.0.2)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.0.4)
blob 1.2.2 2021-07-23 [1] CRAN (R 4.0.4)
broom 0.7.9 2021-07-27 [1] CRAN (R 4.0.4)
BSgenome 1.58.0 2020-10-27 [1] Bioconductor
bslib 0.2.5.1 2021-05-18 [1] CRAN (R 4.0.4)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.4)
callr 3.7.0 2021-04-20 [1] CRAN (R 4.0.4)
car 3.0-11 2021-06-27 [1] CRAN (R 4.0.4)
carData 3.0-4 2020-05-22 [1] CRAN (R 4.0.2)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.0.2)
checkmate 2.0.0 2020-02-06 [1] CRAN (R 4.0.2)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.0.4)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.0.4)
crayon 1.4.1 2021-02-08 [1] CRAN (R 4.0.4)
curl 4.3.2 2021-06-23 [1] CRAN (R 4.0.4)
data.table 1.14.0 2021-02-21 [1] CRAN (R 4.0.4)
DBI 1.1.1 2021-01-15 [1] CRAN (R 4.0.4)
dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.0.4)
DelayedArray 0.16.3 2021-03-24 [1] Bioconductor
DelayedMatrixStats 1.12.3 2021-02-03 [1] Bioconductor
desc 1.3.0 2021-03-05 [1] CRAN (R 4.0.4)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.0.4)
digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.2)
dplyr * 1.0.7 2021-06-18 [1] CRAN (R 4.0.4)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.0.4)
evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.2)
fansi 0.5.0 2021-05-25 [1] CRAN (R 4.0.4)
farver 2.1.0 2021-02-28 [1] CRAN (R 4.0.4)
fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.0.3)
forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.0.4)
foreign 0.8-81 2020-12-22 [2] CRAN (R 4.0.4)
fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
GenomeInfoDb * 1.26.7 2021-04-08 [1] Bioconductor
GenomeInfoDbData 1.2.4 2020-11-10 [1] Bioconductor
GenomicAlignments 1.26.0 2020-10-27 [1] Bioconductor
GenomicFeatures 1.42.3 2021-04-01 [1] Bioconductor
GenomicRanges * 1.42.0 2020-10-27 [1] Bioconductor
ggbeeswarm 0.6.0 2017-08-07 [1] CRAN (R 4.0.2)
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.0.4)
ggpubr * 0.4.0 2020-06-27 [1] CRAN (R 4.0.3)
ggsignif 0.6.2 2021-06-14 [1] CRAN (R 4.0.4)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.0.4)
glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.0.2)
gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.2)
haven 2.4.3 2021-08-04 [1] CRAN (R 4.0.4)
highr 0.9 2021-04-16 [1] CRAN (R 4.0.4)
hms 1.1.0 2021-05-17 [1] CRAN (R 4.0.4)
htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.0.4)
httpuv 1.6.2 2021-08-18 [1] CRAN (R 4.0.4)
httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
IRanges * 2.24.1 2020-12-12 [1] Bioconductor
irlba 2.3.3 2019-02-05 [1] CRAN (R 4.0.2)
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.4)
jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.4)
knitr 1.33 2021-04-24 [1] CRAN (R 4.0.4)
labeling 0.4.2 2020-10-20 [1] CRAN (R 4.0.2)
later 1.3.0 2021-08-18 [1] CRAN (R 4.0.4)
lattice 0.20-41 2020-04-02 [2] CRAN (R 4.0.4)
lifecycle 1.0.0 2021-02-15 [1] CRAN (R 4.0.4)
locfit 1.5-9.4 2020-03-25 [1] CRAN (R 4.0.2)
lubridate 1.7.10 2021-02-26 [1] CRAN (R 4.0.4)
magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.3)
MAST * 1.16.0 2020-10-27 [1] Bioconductor
Matrix 1.3-4 2021-06-01 [1] CRAN (R 4.0.4)
MatrixGenerics * 1.2.1 2021-01-30 [1] Bioconductor
matrixStats * 0.60.0 2021-07-26 [1] CRAN (R 4.0.4)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.4)
modelr 0.1.8 2020-05-19 [1] CRAN (R 4.0.2)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.2)
openssl 1.4.4 2021-04-30 [1] CRAN (R 4.0.4)
openxlsx 4.2.4 2021-06-16 [1] CRAN (R 4.0.4)
pillar 1.6.2 2021-07-29 [1] CRAN (R 4.0.4)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.4)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
pkgload 1.2.1 2021-04-06 [1] CRAN (R 4.0.4)
plyr 1.8.6 2020-03-03 [1] CRAN (R 4.0.2)
prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.2)
processx 3.5.2 2021-04-30 [1] CRAN (R 4.0.4)
progress 1.2.2 2019-05-16 [1] CRAN (R 4.0.2)
promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.0.4)
ps 1.6.0 2021-02-28 [1] CRAN (R 4.0.4)
purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.0.4)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.0.4)
RColorBrewer * 1.1-2 2014-12-07 [1] CRAN (R 4.0.2)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.0.4)
RCurl 1.98-1.4 2021-08-17 [1] CRAN (R 4.0.4)
readr * 2.0.1 2021-08-10 [1] CRAN (R 4.0.4)
readxl 1.3.1 2019-03-13 [1] CRAN (R 4.0.2)
remotes 2.4.0 2021-06-02 [1] CRAN (R 4.0.4)
reprex 2.0.1 2021-08-05 [1] CRAN (R 4.0.4)
reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.0.2)
rio 0.5.27 2021-06-21 [1] CRAN (R 4.0.4)
rlang 0.4.11 2021-04-30 [1] CRAN (R 4.0.4)
rmarkdown 2.10 2021-08-06 [1] CRAN (R 4.0.4)
rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.3)
Rsamtools * 2.6.0 2020-10-27 [1] Bioconductor
RSQLite 2.2.8 2021-08-21 [1] CRAN (R 4.0.4)
rstatix 0.7.0 2021-02-13 [1] CRAN (R 4.0.4)
rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.3)
rsvd 1.0.5 2021-04-16 [1] CRAN (R 4.0.4)
rtracklayer 1.50.0 2020-10-27 [1] Bioconductor
rvest 1.0.1 2021-07-26 [1] CRAN (R 4.0.4)
S4Vectors * 0.28.1 2020-12-09 [1] Bioconductor
sass 0.4.0 2021-05-12 [1] CRAN (R 4.0.4)
scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.2)
scater * 1.18.6 2021-02-26 [1] Bioconductor
scuttle 1.0.4 2020-12-17 [1] Bioconductor
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
SingleCellExperiment * 1.12.0 2020-10-27 [1] Bioconductor
sparseMatrixStats 1.2.1 2021-02-02 [1] Bioconductor
splatter * 1.17.2 2021-09-20 [1] Bioconductor
stringi 1.7.3 2021-07-16 [1] CRAN (R 4.0.4)
stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
SummarizedExperiment * 1.20.0 2020-10-27 [1] Bioconductor
testthat 3.0.4 2021-07-01 [1] CRAN (R 4.0.4)
tibble * 3.1.3 2021-07-23 [1] CRAN (R 4.0.4)
tidyr * 1.1.3 2021-03-03 [1] CRAN (R 4.0.4)
tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.0.4)
tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 4.0.4)
tzdb 0.1.2 2021-07-20 [1] CRAN (R 4.0.4)
usethis 2.0.1 2021-02-10 [1] CRAN (R 4.0.4)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.4)
VariantAnnotation * 1.36.0 2020-10-27 [1] Bioconductor
vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.0.4)
vipor 0.4.5 2017-03-22 [1] CRAN (R 4.0.2)
viridis 0.6.1 2021-05-11 [1] CRAN (R 4.0.4)
viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.0.4)
whisker 0.4 2019-08-28 [1] CRAN (R 4.0.2)
withr 2.4.2 2021-04-18 [1] CRAN (R 4.0.4)
workflowr 1.6.2 2020-04-30 [1] CRAN (R 4.0.2)
xfun 0.25 2021-08-06 [1] CRAN (R 4.0.4)
XML 3.99-0.7 2021-08-17 [1] CRAN (R 4.0.4)
xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.2)
XVector * 0.30.0 2020-10-27 [1] Bioconductor
yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
zip 2.2.0 2021-05-31 [1] CRAN (R 4.0.4)
zlibbioc 1.36.0 2020-10-27 [1] Bioconductor
[1] /mnt/mcfiles/cazodi/R/x86_64-pc-linux-gnu-library/4.0
[2] /opt/R/4.0.4/lib/R/library