15 Integrating single-cell ’omics datasets
This is a very big topic, too big to cover in depth in this workshop!
However, we have already seen a few approaches to integrating single-cell RNA-seq data in various ways. This chapter provides the opportunity to discuss these ideas further and develop something of a taxonomy of data integration aims and approaches for single-cell ’omics data.
“Data integration” of single-cell ’omics data may cover any or all of the following:
- Batch correction within an experiment/study;
- Using a reference (“atlas”) to inform analysis of a generated dataset;
- Combining data from the same biological system but across different studies/labs/platforms;
- Making use of multiple ’omics “views”: from the same cells or not;
- … and many more possibilities!
The 12 Grand Challenges in Single-Cell Data Science preprint provides a more systematic way of thinking about the different types of single-cell ’omics data integration that we may want to achieve. The figure below lays out several distinct types of approach.
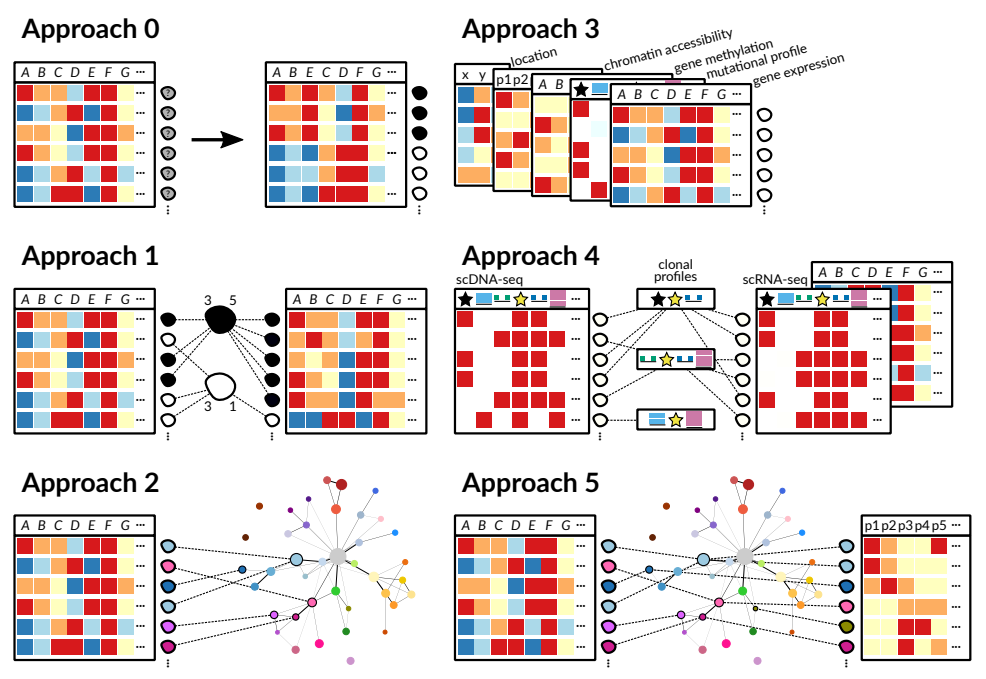
Figure 15.1: Reproduction of Figure 6 from Laehnemann et al (2019). Approaches for integrating single-cell measurement datasets across measurement types, samples and experiments. Approach 0: Clustering of cells from one sample from one experiment, no data integration is needed. Approach 1: Cell populations / clusters from multiple samples but the same measurement type need to be linked. Approach 2: For cell populations / clusters across multiple experiments, stable reference systems like cell atlases are needed (compare Figure 1). Approach 3: Whenever multiple measurement types can be obtained from the same cell, they are automatically linked. However, this setup highlights the problem of data sparsity of all available measurement types and the dependency of measurement types that needs to be accounted for. Approach 4: When multiple measurement types cannot be obtained from the same cell, a solution is to obtain them from cells of the same cell population. However, this combines the problems of Approach 1 with those of Approach 3. Approach 5: One possibility for easing data integration across measurement types from separate cells would be to have a stable reference (cell atlas) across multiple measurement types. Effectively, this combines the problems of Approaches 2, 3 and 4.
This table provides some more details and examples:
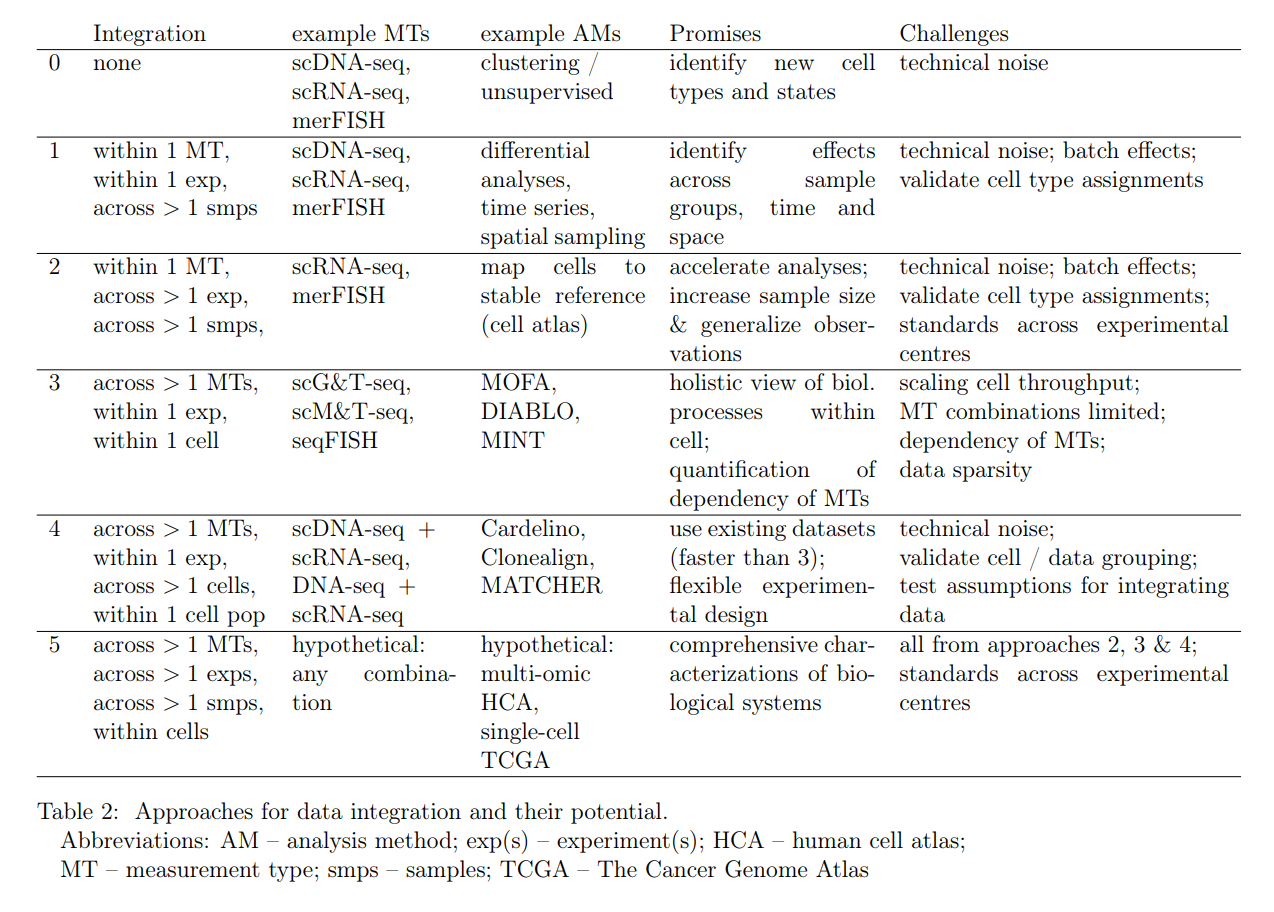
Figure 2.3: Reproduction of Table 2 from Laehnemann et al (2019).
As we can see here, there are many different approaches to integrating data, and the approaches depend on the data types we have and what we want to achieve. Some types of data integration are already eminently feasible; others require much more methods and software development before they are achievable. Ultimately, it all comes back to our biological questions. What questions we want to answer will drive the data we generate and the approaches we might sensibly take to integrate that data.
A final thought: in some (many?) what we might call data synthesis might be preferable to data integration. That is, we might not need or want to combine disparate data sets and data types into one holistic (and likely very challenging) analysis. Rather, we might instead analyse different data sets/types separately and synthesise what we learn from each of them to answer biological questions of interest. Kind of how science is supposed to work!
15.1 Further reading
We recommend that you read section 6.1 of the “Grand Challenges” paper for more detailed discussion of the current status of data integration for single-cell ’omics data and open problems that remain:
- Laehnemann,D. et al. (2019) 12 Grand challenges in single-cell data science PeerJ Preprints. link